https://ift.tt/LMsCYkl Photo by Philip Oroni on Unsplash Introduction Pandas is one of the most popular data science libraries. From...

Introduction
Pandas is one of the most popular data science libraries. From the official documentation:
pandas is a fast, powerful, flexible and easy-to-use open-source data analysis and manipulation tool, built on top of the Python programming language
And everyone who is familiar with pandas will confirm these words with confidence. And at the same time, many in their pipelines are faced with the fact that pandas does not use the full power of the CPU. In other words, pandas methods do not support concurrency.
Consider a simple example:
synchronous describe time took: 37.6 s.
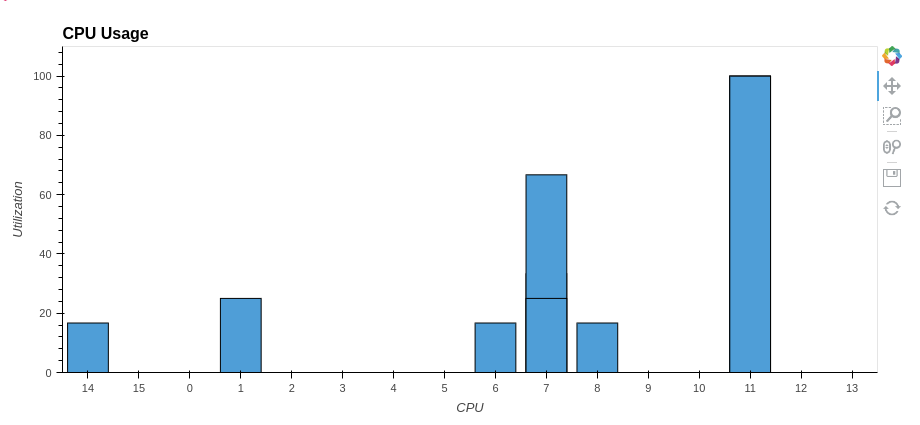
As you can see from the graph above, only one core was used at 100%. The rest were idle. But what if you use all the processor cores? How quickly can you get the same result and most importantly — how to achieve this?
So let’s first think about how describe() works? it processes the columns and calculates some statistics for each. We have a dataframe with 1000 columns. And what about dividing our dataframe along the first axis (along the columns) into several smaller parts, separately calculating describe for each part, and then combining the result back into one common dataframe? Let’s see how we can split the dataframe into parts along the specified axis. This can easily be done with the array_splitfunction of the numpy module. Let’s look at a simple example:
[ cat dog
0 0 0
1 1 1
2 2 2,
cat dog
3 3 3
4 4 4
5 5 5,
cat dog
6 6 6
7 7 7,
cat dog
8 8 8
9 9 9]
And split by columns
[ cat
0 0
1 1
2 2
3 3
4 4
5 5
6 6
7 7
8 8
9 9,
dog
0 0
1 1
2 2
3 3
4 4
5 5
6 6
7 7
8 8
9 9]
So, we have an algorithm for splitting a large dataframe into smaller parts along the axis we need. What’s next? How to make all the cores of our CPU work? As you know, pandas under the hood uses numpy arrays and methods for working with them, which often removes the global interpreter lock (GIL). Thus, we can use the thread pool to parallelize calculations. Let’s turn to the map method of the ThreadPoolExecutor class of the concurrent.futures module:
map(func, *iterables, timeout=None, chunksize=1)
Similar to map(func, *iterables) except:
1. the iterables are collected immediately rather than lazily;
2. func is executed asynchronously and several calls to func may be made concurrently.
In our case, the describe() method is the function, and the iterable object is a list of dataframes. The full code with the implementation of the pipeline is given below.
parallel desribe time took: 15.0 s.
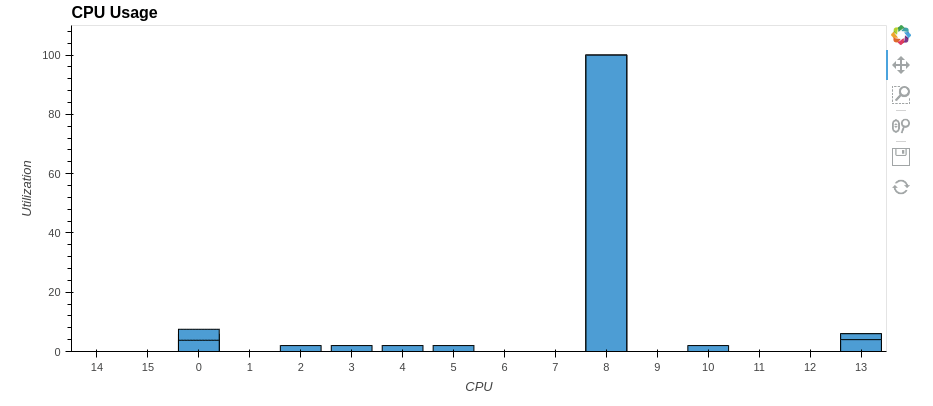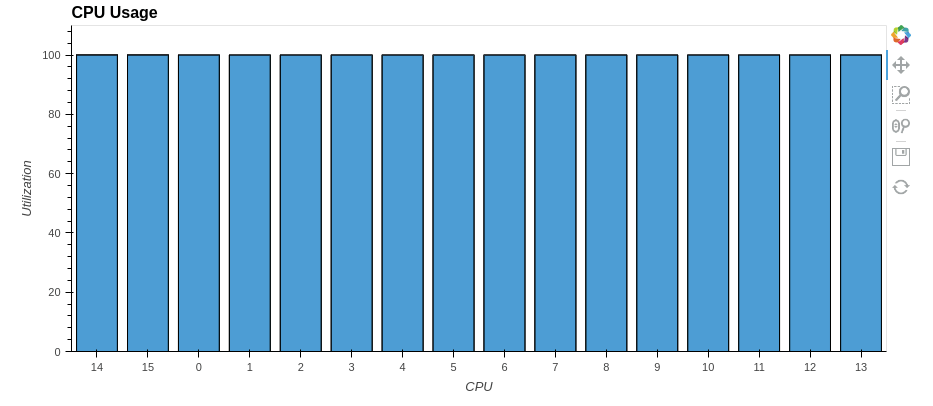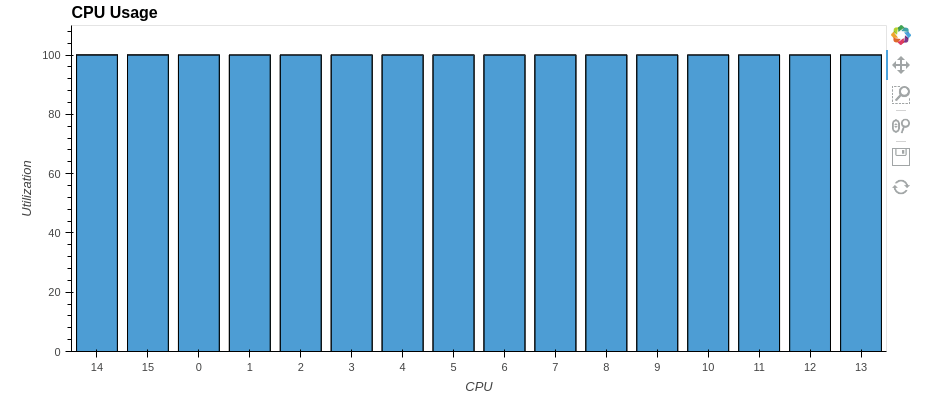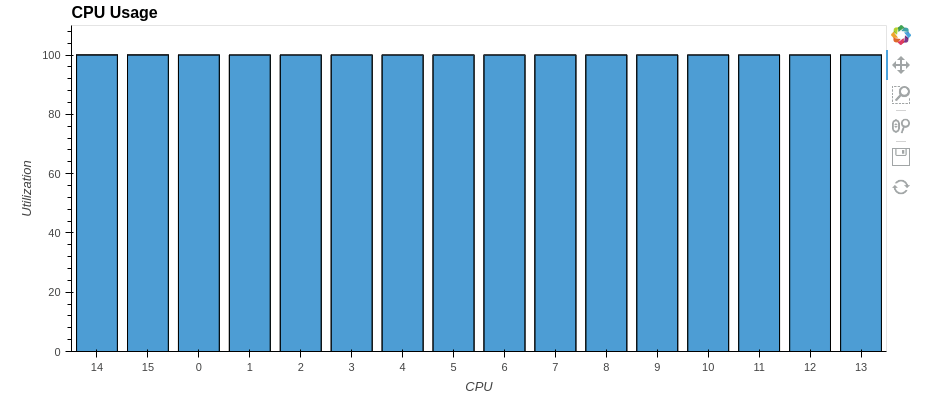
So we’ve sped up the pandas.DataFrame.describe() method by a factor of two! Similarly, you can parallelize other pandas methods.
Introduction to parallel-pandas
The parallel-pandas library locally implements the approach to parallelizing pandasmethods described above. First, install parallel-pandasusing the pip package manager:
pip install --upgrade parallel-pandas
Just two lines are enough to get started!
synchronous time took: 3.7 s.
parallel time took: 0.688 s.
As you can see the p_quantile method is 5 times faster!
Once parallel-pandas is initialized, you continue to use pandas as usual. Parallel analogs of pandas methods are prefixed with p_ from the word parallel. Moreover, parallel-pandas allows you to see the progress of parallel computations. To do this, it is enough to specify disable_pr_bar=False during initialization.
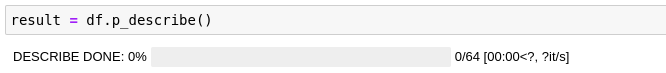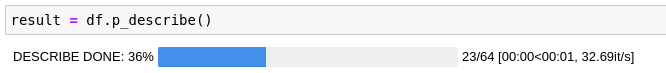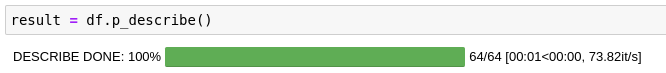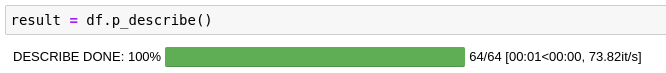
As you can see, parallel-pandas takes care of splitting the original dataframe into chunks, parallelizing and aggregating the final result for you.
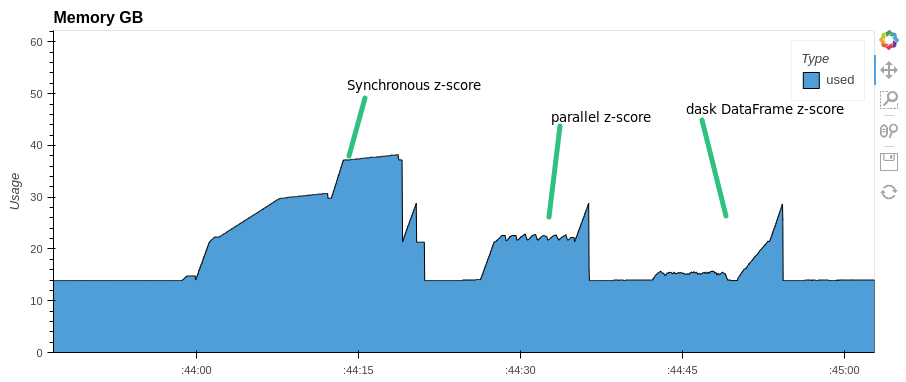
More complex calculations can be parallelized in a similar way. For example, let’s z-normalize the columns of our dataframe. Also, for comparison, we implement z-normalization with dask dataframe:
synchronous z-score normalization time took: 21.7 s.
parallel z-score normalization time took: 11.7 s.
dask parallel z-score normalization time took: 12.5 s.
Pay attention to memory consumption. parallel-pandas and dask use almost half as much RAM as pandas
For some methods parallel-pandas is faster than dask DataFrame:
dask nunique time took:42.5 s.
dask rolling window mean time took: 19.8 s.
paralle-pandas nunique time took:12.5 s.
parallel-pandas rolling window mean time took: 11.8 s.
parallel-pandas implements many pandas methods. The full list can be found in the documentation.
To summarize:
- in the article, we briefly reviewed the idea of parallelizing pandas methods and implemented it using the describe method as an example
- We also got acquainted with the parallel-pandas library which makes it easy to parallelize the basic pandas methods.
- We also compared the work of parallel-pandas with another well-known library for distributed computing dask using several examples.
I hope this article helps you, good luck!
Easily parallelize your calculations in pandas with parallel-pandas was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
from Towards Data Science - Medium
https://towardsdatascience.com/easily-parallelize-your-calculations-in-pandas-with-parallel-pandas-dc194b82d82f?source=rss----7f60cf5620c9---4
via RiYo Analytics

ليست هناك تعليقات