https://ift.tt/kCYGHQp Strategies on how to throttle the data retrieval rate using PubChem chemical safety data as a use case Photo taken...
Strategies on how to throttle the data retrieval rate using PubChem chemical safety data as a use case

Table of contents
· Setting the scene
· Synchronous data retrieval
· Asynchronous data retrieval
∘ Conceptualise rate throttling
∘ Concurrent and rate-throttled retrieval using asyncio
∘ Overview of results
· Conclusions
Setting the scene
As a general rule, orchestrating I/O-bound processes can be achieved using a single core by cleverly switching context whilst the external resource is taking its time to respond. Still, there are several approaches that can be used and differ on two main aspects:
- whether they use preemptive or cooperative (also known as non-preemptive) multitasking that has to do with who controls the context switch
- how data retrieval can be throttled to meet the requirements of the data provider
The latter is in my view the most important. When interacting with data providers we need to ensure that the data retrieval rate does not exceed the limit specified by the service we are using. It is the responsibility of the data analyst to act responsibly and not only focus on optimising the process from the data recipient point of view.
This article uses chemical safety as a theme. It collects chemical safety data from PubChem for all industrial chemicals used in European Union using cooperative multitasking based on asyncio. In European Union substances that are manufactured or imported in a quantity of 1 t/y or more need to be registered according to the requirements of the REACH Regulation. The list of registered substances can be conveniently downloaded from here as a nicely formatted excel file that looks like below (Figure 1). Even if this list is not available in the future any list of substance identifiers can be used as input. Alternatively the list used when this article was written can be obtained from the accompanying repository.
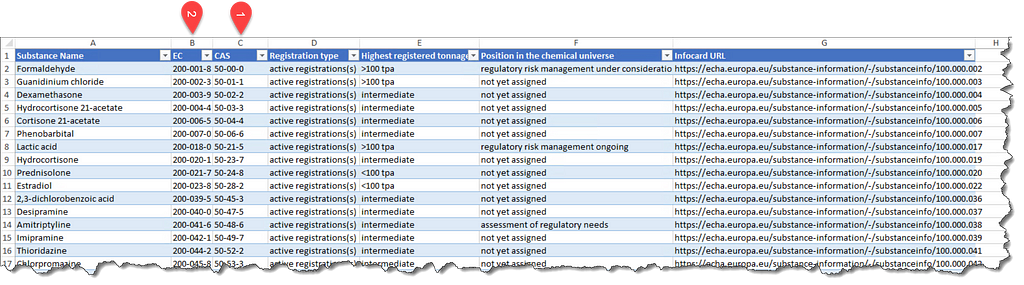
The list of substances can be read using pandas with the code below.
There is nothing special here. In case the filtering is not clear, the intention is to keep the substances that have a CAS number, that is an identifier that can be readily used to retrieve data from PubChem. Other identifiers can be used too with little additional effort as explained in the PubChem PUG REST documentation and tutorial.
The use case of this article involves two steps. The first step is to use the CAS number to obtain the corresponding PubChem compound ID (CID). As an example, formaldehyde has the CAS number 50–00–0, that can be used to obtain the corresponding CID using a POST request
https://pubchem.ncbi.nlm.nih.gov/rest/pug/compound/name/cids/JSON
with the header
Content-Type: application/x-www-form-urlencoded
and body
name=50-00-0
At the time of writing this gives the CID 712. It is possible that the CAS number can correspond to more than one CID that is handled by the implementation. This step can also be accomplished by using a simpler GET request, but we prefer the POST functionality offered by PubChem because it does not suffer from the limitations of the characters that can be passed in the URL (e.g. when obtaining the CID from the substance name). This is less important when the starting point is a CAS number but I opted for a more generalised implementation.
The second step is to obtain the index for the particular compound, i.e. a listing of what information is present, but without the entire data content; essentially a table of contents for that record. In the case of formaldehyde this can be obtained using a GET request
https://pubchem.ncbi.nlm.nih.gov/rest/pug_view/index/compound/712/JSON
The use case essentially allows analysing the type of information that is available in PubChem for industrial chemicals. PubChem is an expansive repository that provides comprehensive chemical information and fluent programmatic access built for data-intensive research. This article uses only part of the offered functionality to explain concurrency in Python that can be extended further to obtain more data from PubChem, such as bioassay data. Still, the use case presented is sufficiently complex to be representative of the challenges that such integration may ensue.
Synchronous data retrieval
Before thinking about concurrency it is useful to test the data retrieval approach using a synchronous (blocking) approach based on the excellent requests package.
The first step related to the retrieval of the CID(s) from the CAS number is implemented with the function retrieve_CID, whilst the second step related to the retrieval of the PUG VIEW index for a particular CID is implemented with the function retrieve_pugview.
The approach uses CAS numbers as a starting point. However, the implementation also allows using the EC number or substance name as starting point in case readers find this useful. Due to this there is a need for URL and byte encoding that are not explained in full detail, but are hopefully clear from the code. Similarly, I do not provide an explanation for the import logger import and the logging approach in general as this is not important for this article. For a refresher on logging please refer to this excellent resource. The synchronous approach can be further optimised by retrying failed attempts and by creating a requests session, but this was not pursued in the synchronous functions as the focus of this articles is the asynchronous implementation. The complete code can be seen in the accompanying GitHub repository.
Coming back to the formaldehyde example, we can retrieve data synchronously with
that returns the same JSON output we can see by using the CID for formaldehyde in the browser
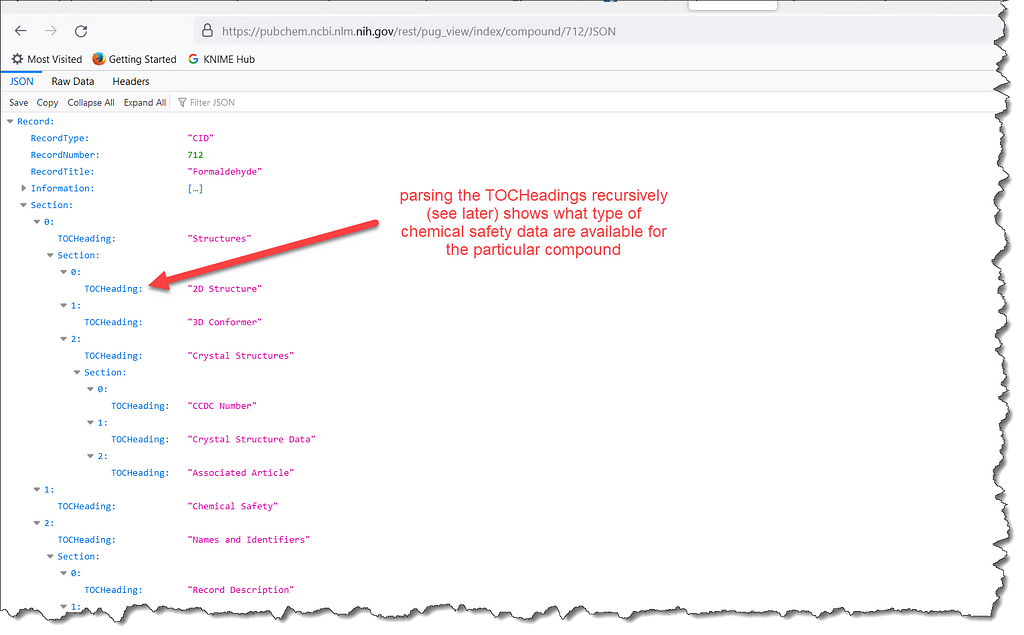
A particularly interesting section of the response is the nested TOCHeading values that can be parsed recursively to see how much data PubChem contains when it comes to industrial chemicals in European Union. We will come back to this towards the end of the article.
This synchronous approach is perhaps the simplest way to obtain data programmatically from PubChem and may already be useful to some readers. However, the purpose of this article is to go beyond the synchronous approach and show how the two REST requests can be executed concurrently for a fairly large number of CAS numbers (but not so large to think about the ftp data retrieval option offered by PubChem). Concurrency is not the easiest topic to wade into. Hence, before working concurrently it is always advisable to implement a synchronous approach. This ensures that the REST API is used correctly and sets a baseline to check any performance gains of the concurrent approach. For the record, retrieving the index of formaldehyde starting from its CAS number takes approximately 1.3 sec.
Asynchronous data retrieval
Conceptualise rate throttling
One of the most important aspects when using APIs for retrieving data is respecting the terms and conditions of the data provider that may impose restrictions on what can be done with the retrieved data. In our case there are no such complexities because the data will not be used for any particular application as the intention is to demonstrate the process for retrieving the data rather than building a user facing application or exploiting the data in a different way. In addition to the data usage it is important to responsibly retrieve data so that we do not put excessive strain on the data provider’s servers. This section attempts how to approach rate throttling for our use case.
PubChem imposes request volume limitations that require programmatic web requests to be throttled so that there are:
- no more than 5 requests per second
- no more than 400 requests per minute
- no longer than 300 second running time per minute
The second requirement seems redundant given that if we submit 5 requests per second for the whole minute then the number of requests submitted in one minute will be 300, i.e. less than 400. Which condition may be the limiting one depends on the response time for a typical request, that in turn may depend on the volume of the requested data, the overall load of the service and nuances related to PubChem’s load balancer. All in all, this means that we cannot assume that the same request will take the same time if executed at different times. The approach should be generalised and flexible to account for such fluctuations.
The starting point for orchestrating the data retrieval is that we can submit up to five requests per sec. If we assume that we are running five separate and concurrent streams then all requirements can be met if the response time of each request is measured so that:
- a response that takes t < 1 sec is followed by 1-t sec idle time
- a response that takes t ≥ 1 sec can be immediately followed by another request
Four different situations are simulated below (the repository contains the function visualise_task_orchstrationinin utilities.py in case of interest) differing in the response time distribution. We assume five concurrent streams each issuing ten requests. In Figure 2 the response time is fairly short and follows a normal distribution with mean 0.2 sec and standard deviation 0.05 sec. We can see that the rate limiting condition is the number of requests per second. The five concurrent streams submit collectively 5 requests per second, but the response time (load on PubChem server) is only 20% of the wall time, whilst 80% is idle time. This means that we only use 60 sec running time per minute but unfortunately we cannot push it and any further without violating the request volume limitations.

Figure 3 simulates slightly longer response times, by drawing from a normal distribution with mean 0.5 sec and standard deviation 0.12 sec. Once more the rate limiting condition is the number of requests per second. The running time is now roughly 5x0.5x60 = 150 sec per minute.
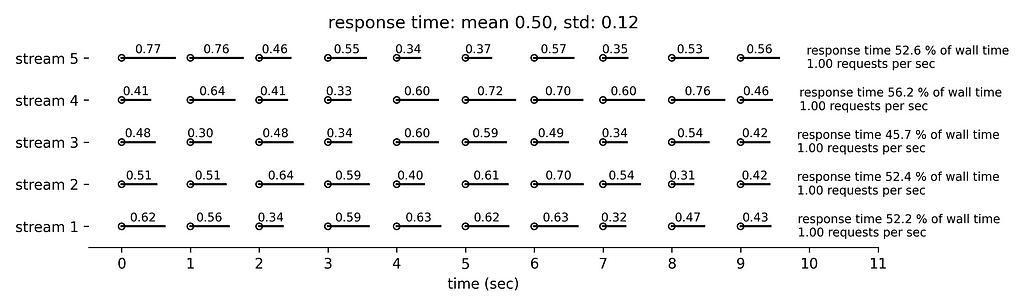
Figure 4 simulates even longer response times with mean 1 sec and standard deviation 0.25 sec. We can see that the five concurrent streams submit collectively slightly less than 5 requests per second and the running time is slightly less than 300 sec per minute.
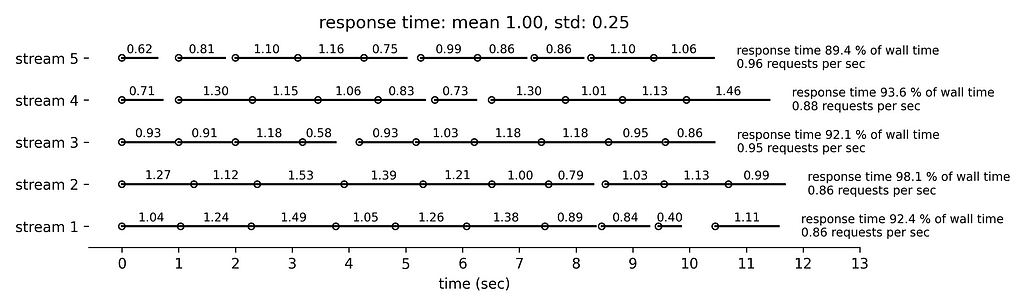
Finally, Figure 5 simulates the longest response time with mean 1.5 sec and standard deviation 0.38 sec. Very few requests are followed by idle time. The rate limiting condition is the running time that approaches 300 sec per min.
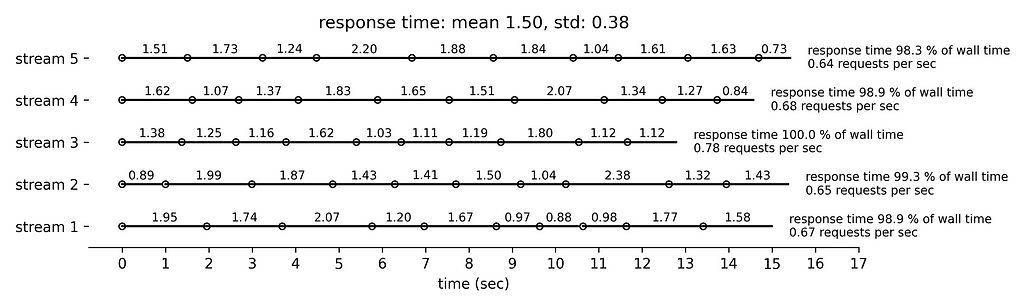
In practice, and because of the complexity of the request life cycle and the challenges in measuring accurately the response time, we will impose a slightly longer idle time by 0.1 sec to allow for some error margin.
The above approach can be improved further especially if the response times fluctuate, e.g. by following a binomial distribution with faster and slower responses. In this case, the faster responses could be followed by less idle time because the slower responses ensure that we will not be reaching the five requests per sec limit so easily. However, such approach is more complex to develop and potentially dangerous if left running unsupervised, because unless we can theoretically guarantee that the request volume limitations cannot be exceeded then we can inadvertently stress PubChem and eventually cause out IP to be blocked.
We note that PubChem also offers dynamic throttling. The HTTP response headers accompanying all PUG-REST web requests contain information on how close the user is to approaching the limits. The limits can be dynamically adjusted by PubChem based on the overall demand and hence can be used in real time to ensure that the concurrency rate is dynamically adjusted accordingly. This feedback loop mechanism goes beyond the scope of this article that has to cover quite some ground. In this article dynamic throttling is only used to double check that the data retrieval rate remains within the acceptable boundaries.
In the following section we will implement the data retrieval approach using five independent and concurrent streams to retrieve the CID(s) for several thousand substances using their CAS numbers as a starting point. These CIDs will be made unique and then used to retrieve their corresponding PUG VIEW indices.
Concurrent and rate-throttled retrieval using asyncio
The asynchronous approach is heavily based on the synchronous one but with several tweaks listed in this session.
The most critical tweak is the replacement of the blocking requests module by the aiohttp module, which is a library to make HTTP requests in an asynchronous fashion. The requests themselves are made using a single aiohttp session, to exploit the reusage of the session’s internal connection pool.
Although rare, the implementation resubmits requests that may temporarily fail, e.g. due to network issues or because PubChem’s servers are busy. Looking at the logs this only happens a few tens of times and no request had to be submitted more than twice.
Rate throttling is achieved via an asyncio semaphore defined with
sem = asyncio.Semaphore(limit)
with limit=5. A semaphore manages an internal counter which is decremented by each sem.aquire() call (once we enter the context manager using the semaphore) and incremented by each sem.release() call (once we exit the context manager using the semaphore). The counter can never go below zero; when sem.aquire() finds that it is zero, it blocks, waiting until some task calls sem.release()that wakes up another task waiting to acquire the semaphore. This essentially implements the five concurrent and independent streams conceptualised earlier.
Short requests are followed by an asyncio.sleep(idle_time) call where the idle time depends on the request duration that is assumed to reflect the PubChem computing time. Luckily, aiohttp supports client tracing that works by attaching listeners coroutines to the signals provided by the TraceConfig instance:
async def on_request_start(session, trace_config_ctx, params):
trace_config_ctx.start = asyncio.get_event_loop().time()
async def on_request_end(session, trace_config_ctx, params):
elapsed_time = asyncio.get_event_loop().time() - trace_config_ctx.start
if trace_config_ctx.trace_request_ctx['request duration'] is not None:
raise Exception('should not happen')
trace_config_ctx.trace_request_ctx['request duration'] = elapsed_time
trace_config = aiohttp.TraceConfig()
trace_config.on_request_start.append(on_request_start)
trace_config.on_request_end.append(on_request_end)
This instance is used as a parameter for the ClientSession constructor having as a result a client that triggers the different signals supported by TraceConfig. The signals take as first argument the ClientSession and as a second a SimpleNamespace called trace_config_ctx, that can be used to share the state through to the different signals that belong to the same request and to the same TraceConfig class. This enables measuring the elapsed time from the start to the end of the request. The trace_config_ctx param is initialized at the beginning of the request flow. An added complexity is that we need to pass information from the listener back to the coroutine that implements the request so that the idle time can be set. The code makes uses of the trace_request_ctx param that be can given at the beginning of the request execution, accepted by all of the HTTP verbs, and will be passed as a keyword argument to the default factory that instantiates trace_config_ctx. We are not using this functionality to pass data to the listener but to obtain the request duration, as the value of the key value pair trace_request_ctx = {'request duration': None} will be set once the request returns.
Admittedly this section has introduced a lot of concepts. It may be easier to look at the whole implementation in the accompanying repository.
Overview of results
The number of substances with a CAS number is 17 829, that correspond to 17 700 unique CAS numbers. Using the coroutine retrieve_CID we obtain 15 361 unique CID numbers with 12 873 CAS numbers having only one CID number. 4 117 CAS numbers are not found in the PubChem database and 710 CAS numbers have two or more CID numbers. The latter are indicative of the substance identification challenges of industrial chemicals and potential duplicate records in PubChem. All in all, the whole data retrieval took 2.04 hours that is by no means prohibitive even if the data retrieval needs to be periodically repeated. Processing the logs, we can see that the average response time is a bit less than 0.2 sec for both request types and the spread of response times is fairly narrow as can be seen below. Still, there is a number of outliers. The code to produce the boxplot using seaborn can be also be found in supplementary repository.
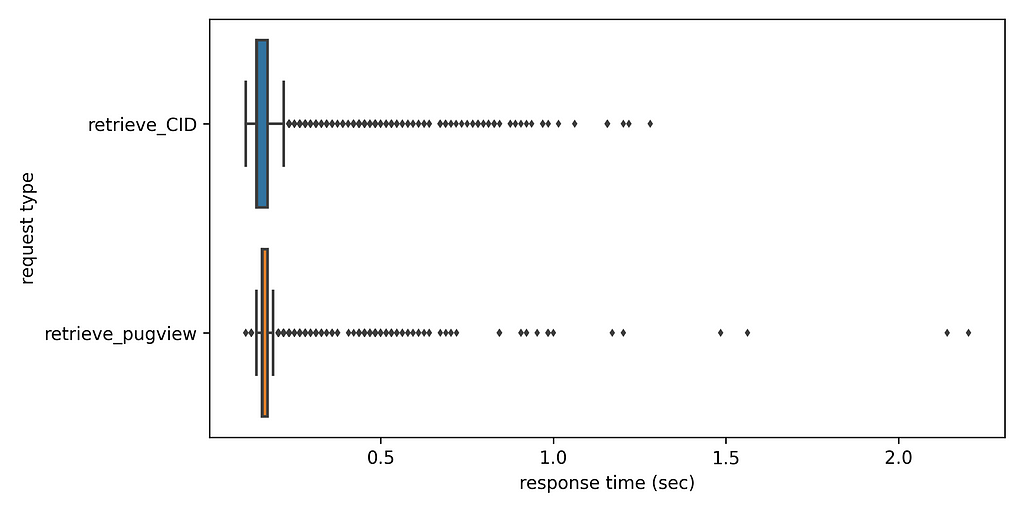
Examining the logs there are ~30 warnings related to exceedance of PubChem’s request volume limitations with response code 503 and message “Too many requests or server too busy”, that is fairly small compared to the ~33k requests. This is a fairly small number and in the end the data retrieval was successful upon retry. The reasons why the throttling led to this are probably not worth examining, given the difficulties in interpreting precisely what PubChem means with computing time and what exactly our client tracing approach measures as response time. PubChem allows for some leeway that we are clearly using responsibly.
Using the recursive function utilities/parse_TOCHeadingsin utilities.py we see that PubChem contains bioassay data for 6 248 CID numbers, i.e. for almost half of the retrieved CID numbers. Such data can be useful in hazard assessment, whilst PubChem also contains significant volume of use and exposure information that can be valuable in risk assessment. The repository contains the full data availability of all TOC headings in this file.
Conclusions
There are other ways to implement rate-throttling concurrent data retrieval, e.g. by combining asyncio with a queue. aiohttpalso allows limiting the number of simultaneous connections but I decided to follow the semaphore route because it is a more general throttling solution that only depends on asyncio itself. Data retrieval is not the only part that can be executed asynchronously. For example, the retrieved data could be stored asynchronously in a PostgreSQL database or in the file system. The list of asyncio libraries and frameworks is constantly growing and many excellent resources are now available (for an example please read this excellent article and browse through the resource links at the end). In fact, there are even alternatives to asyncio, such as curio and trio. I do not know enough on the maturity level of these alternatives. I do recognise that asyncio has evolved rapidly and it is not so easy to understand when to use things like futures, tasks, and coroutines, but at the same time it is included in the standard library and it has now sufficient momentum. Concurrency is not an easy topic and imagination has no limits. For an example, it is possible to run multiple asyncio loops in different cores that may concurrently melt your laptop and brain (pun intended). Jokes aside, I feel that asyncio is sufficient for most IO-bound processes typically encountered by data analysts and data scientists.
For the sake of completeness, IO-bound processes can also be orchestrated using multithreading. My personal view is that asyncio is a better choice than multithreading because switching context is explicit and there is no need to worry about thread safety, without this implying that asyncio does not have its own challenges.
The implementation is this article used high level asyncio functionality, such as asyncio.gather(). Interacting with the loop and the tasks more explicitly can lead to further optimisations so that data retrieval operates even closer to the limit of PubChem’s request volume constraints. Whether this is worth pursuing is a matter of personal taste given that performance gains will likely be marginal. Moreover, the implementation will inevitably become more involved and asyncio is not only complex but constantly evolving. Maintaining the code requires quite some mental capacity to read and experiment with newer features and this may outweigh the benefits, unless if you are theoretically inclined and enjoy the challenge more than you need the results. People have different perspectives and worldviews and if you are up to the challenge please drop me a comment pointing to your approach. I will be more than happy to read more on it!
Disclaimers
The substance list obtained from European Chemicals Agency is subject to the legal terms and conditions as explained here and in the disclaimer. The PubChem data usage policies and disclaimers can be found here. Please consult these pages prior to making use of any of the open data discussed in this article.
Responsible Concurrent Data Retrieval was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
from Towards Data Science - Medium https://ift.tt/F8LECyT
via RiYo Analytics

ليست هناك تعليقات