https://ift.tt/oErBt8U I explain how I utilised the ULMFiT approach in a step-by-step manner in this NLP task to predict medical specialit...
I explain how I utilised the ULMFiT approach in a step-by-step manner in this NLP task to predict medical specialities from patients’ transcripts

Introduction
The problem of predicting one’s illnesses wrongly through self-diagnosis in medicine is very real. In a report by the Telegraph, nearly one in four self-diagnose instead of visiting the doctor. Out of those who misdiagnose, nearly half have misdiagnosed their illness wrongly. While there could be multiple root causes to this problem, this could stem from a general unwillingness and inability to seek professional help.
11% of the respondents surveyed, for example, could not find an appointment in time. This means that crucial time is lost during the screening phase of a medical treatment, and early diagnosis which could have resulted in illnesses treated earlier was not achieved.
With the knowledge of which medical specialty area to focus on, a patient can receive targeted help much faster through consulting specialist doctors. To alleviate waiting times and predict which area of medical speciality to focus on, we can utilise natural language processing (NLP) to solve this task.
Given any medical transcript or patient condition, this software would predict the medical specialty that the patient should seek help in. Ideally, given a sufficiently comprehensive transcript (and dataset), one would be able to predict exactly which illness he is suffering from.
In this case, we would require a text-classification model to classify what medical speciality the text transcript corresponds to.
In this article, I’ll be covering the few main sections:
- The tech stack
- An introduction to Universal Language Model Fine-Tuning (ULMFiT)
- Creating a language model + What goes in a language model?
- A more in-depth explanation of the ULMFiT approach
- Implementation of the ULMFiT approach into the given task (predicting medical specialties)
- Learning points + Nightingale OS + Future plans
The Tech Stack
Enter the tech stack: natural language processing (NLP) and recurrent neural networks (RNNs). Natural language processing is a subset of self-supervised learning. As defined in fast.ai, self-supervised learning trains a model using labels that are already pre-embedded in the independent variable, rather than requiring external labels. For instance, this would be training a model to predict the next word in a text. That seems like a pretty tough task. Nonetheless, OpenAI researchers have recently fully released GPT-3, a language model that can similarly predict the next word in a text with uncanny accuracy, for developer and enterprise use.
Why would we want to use this technique to aid in a classification task where only the prediction based on the dependent variables are needed? If we think about it logically, we do not need a language model which can predict the next few words, we would just require a text-classifier model. You can find the code to this task (with partial annotations ) here.
A Gentle Introduction to ULMFiT: Drastically Increasing Accuracy
It turns out that this is a form of inductive transfer learning, through using the ULMFiT approach proposed by Jeremy Howard and Sebastian Rudder. It “significantly outperforms the state-of-the-art [as of 2018] on six text-classification tasks, reducing the error by 18–24% on the majority of datasets.” (Howard and Ruder, 2018). Basically, it has been empirically found that this method significantly improves accuracy.
I’ll give a high-level overview of how ULMFiT works before diving into the more technical details of how a language model works in general. The ULMFiT architecture basically follows this line of logic: [Step 1] taking a pre-trained language model (e.g. WikiText 103, which can generate text based on prompt inputs), [Step 2] fine-tuning this pre-trained language model, before saving this pre-trained language model as an encoder and [Step 3] utilising this encoded learner to train a text classifier learner. We will analyse practises to be used within these 3 steps to improve accuracy to a greater extent.
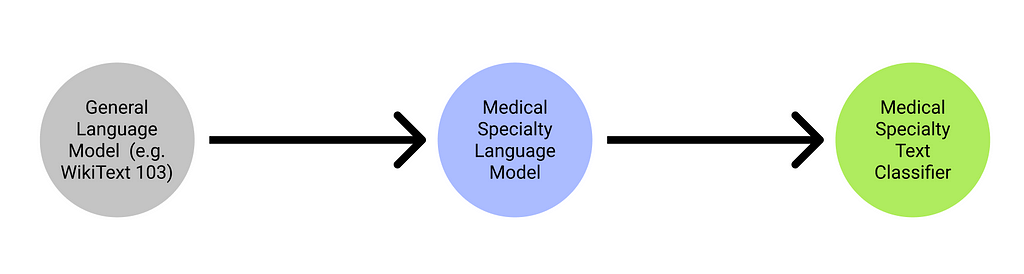
Creating a language model (for step 2)
We first import the data using the kaggle API:
And accordingly unzip the csv file (a single csv file in this case):
As a pre-processing step, we remove several irrelevant rows of information which does not have a legitimate medical speciality attached to the patient transcript:
What goes in a language model?
There are several key steps that are required for creating a language model.
- Tokenization: which is to convert the text into a list of words
- Numericalisation: making a list of all the unique words that appear (a vocab list), and convert each word into a number, by looking its index up in the vocab list
- Loading numericalised tokens into a language model dataloader
The end goal is for our language model to predict the next word in a sentence, (i.e. given the prompt ”My name is”, the model should be able to output a name.).
Understanding tokenization: A token is basically an element of a list created by the tokenization process. It could be a word, part of a word, or a single character. We want to utilise word tokenization for this task, which creates tokens for each word. This would result in some pretty funny outputs, as shown:
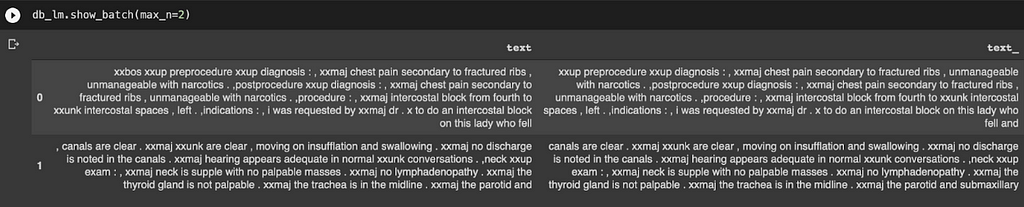
Instead of displaying the text, ‘words’ like “xxbox” and “xxmaj” are produced. What is going on?
It turns out that those special ‘words’ with the character ‘xx’ before them are called special tokens (created by fastai), where part of words which means something significant (a full stop, a capital letter, an exclamation mark, etc.) in English may be hard to interpret by the computer. For instance, “xxmaj” means that the next word begins with a capital, since we have converted all the letters in the entire text file to lowercase.
Understanding numericalisation: We basically need to turn this array of tokens that we see into a tensor, which assigns an index to each unique token. For example, the token “xxmaj” would be one index, and the token “chest” would be another index. Since our model can only receive such a tensor of integers, it is imperative for us to do this.
How then do we go about doing this?
Well, it turns out fastai has a useful shortcut to circumvent such tedious pre-processing. We can use a TextBlock to pass into a DataBlock, before creating a DataLoaders object, as shown:

Following that, we can create a language_model_learner, utilising the AWD_LSTM architecture, and creating a list of vocab that we can utilise later when creating the actual text_classifier_learner. This language_model_learner would be utilising a recurrent neural network (RNN) to train, for a super in-depth discussion you can check out FastAI’s in-depth explanation of RNNs and LSTMs here.
We then train this language_model_learner to improve on the accuracy by trying to predict the next word, before saving it as an encoder, which means that the model does not include the task-specific final layers. In this context, it means that the final output layer of the language model (which predicts and creates words based on prompts) is omitted, allowing us to fine-tune the last layer for our task, which outputs sentiment analysis and classification of prompts into the suitable field of medical specialty.
This makes sense, because we don’t want our text classifier to be predicting words, we want it to be classifying medical specialities based on medical transcripts.
We go about training the language model as follows:
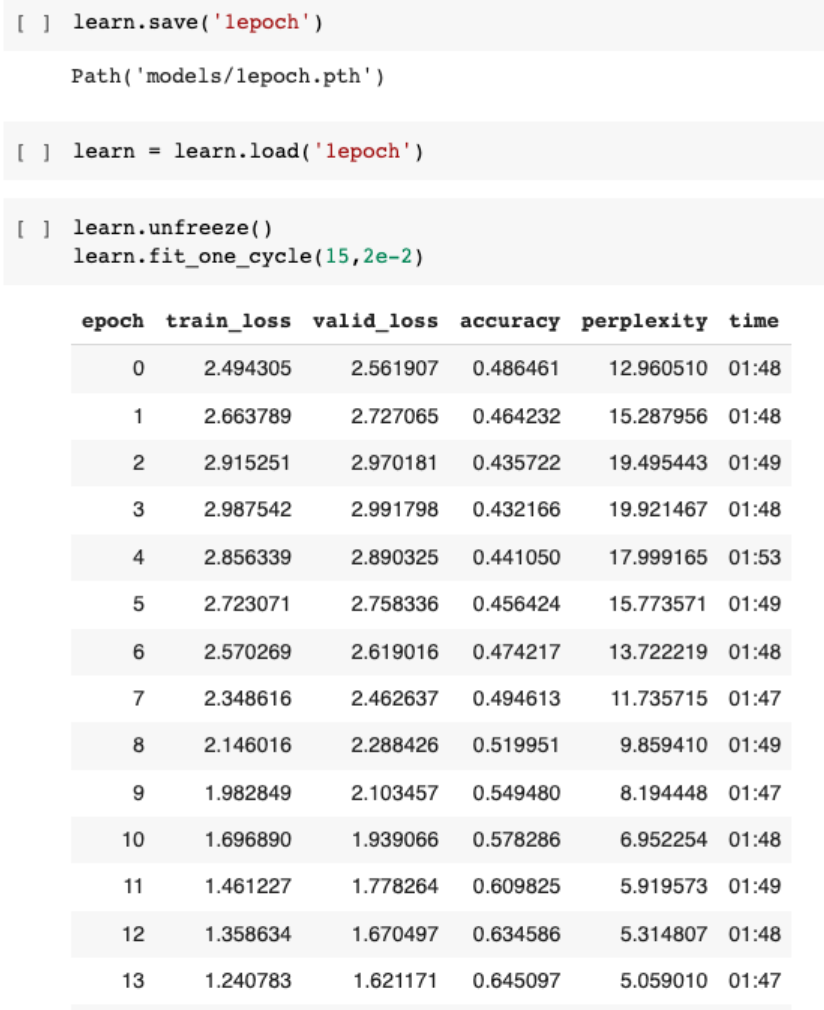
We achieve an accuracy of about 65%, meaning that based on a prompt, our model would be able to predict what the next words are correctly and accurately 65% of the time, based on this dataset. This seems pretty impressive, so let’s get down to work on our actual task.
Before we move on to the techniques used in creating a text-classifier-learner, here’s a more in-depth explanation of the ULMFiT technique.
A Comprehensive Overview of ULMFiT
In Jeremy’s and Sebastian’s breakthrough paper, there are three supplemental practises used in conjunction with ULMFiT:
- Discriminative fine-tuning [used in step 2/3]
- Slanted triangular learning rates [currently unused in this project]
- Gradual unfreezing [used in step 3]
Through combining these practises, they were able to achieve an increase in prediction accuracy for training. Let’s break down these terms piece-by-piece. In the later section of this article, I’ll include my own code to demonstrate how these practises will be implemented in this particular medical transcription task.
Discriminative fine-tuning: Discriminative fine-tuning basically means training different layers of the model at different rates, to account for the different types of information captured, instead of training the entire model at the same learning rate. To this end, [Howard and Rudder] empirically found it to work well to first choose a learning rate as follows:
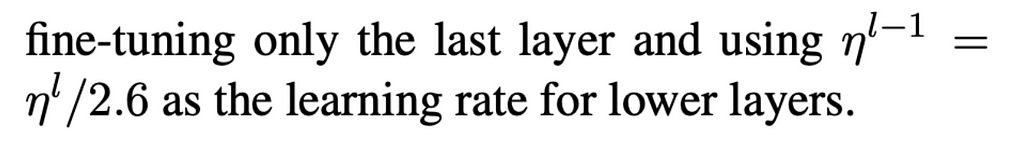
where:
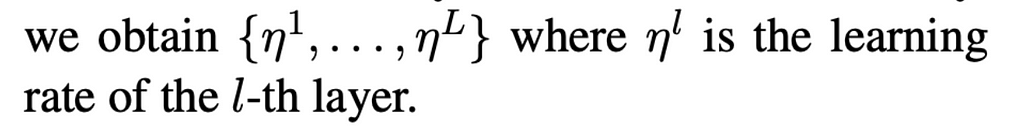
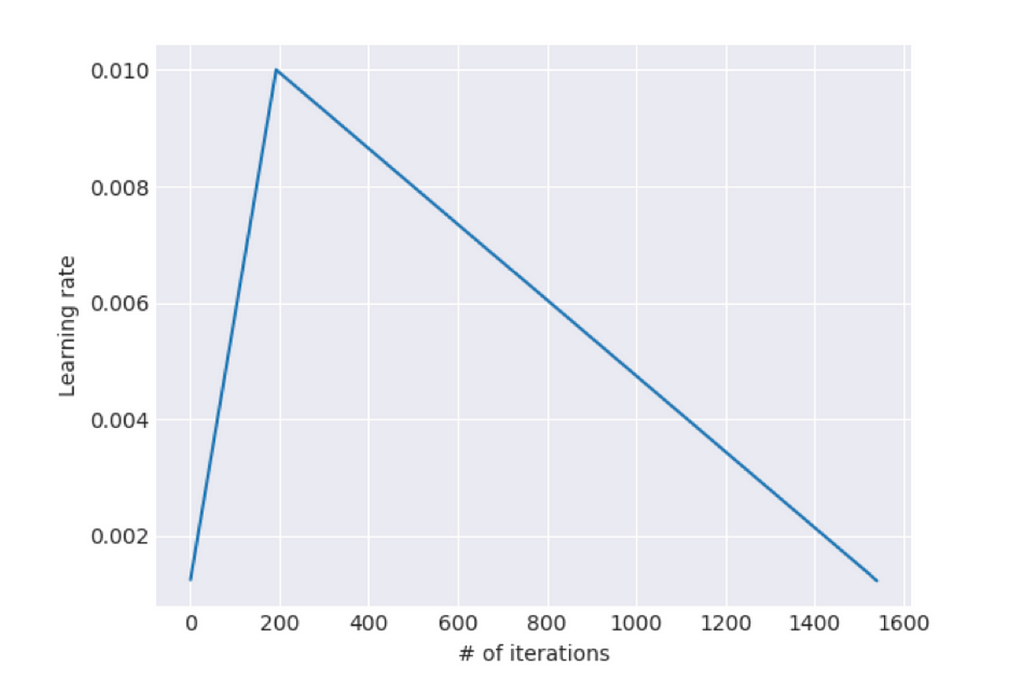
Slanted triangular learning rates: It is defined as a method which “first linearly increases the learning rate and then linearly decays it according to a [mathematical function]”. This ultimately results in a learning rate finder that looks like this:
Gradual unfreezing: To understand unfreezing, we must first comprehend the concept of basic freezing. Freezing basically prevents well-trained weights from being modified by not changing the weights for certain layers. In this case, gradual unfreezing means “first unfreezing the last layer and fine-tuning all unfrozen layers (i.e. fine-tune the last layer) for one epoch, [before] unfreeze[ing] the next lower layer and repeating the process until convergence at the last iteration.”
Through utilising a combination of these 3 techniques, it was empirically found to increase the accuracy of the language model drastically. For discriminative fine-tuning and gradual unfreezing, Jeremy offered a high-level intuition of why these two practises were used. Since the earlier layers of a language model trained to predict would most probably contain weights that would be used in the same fashion as sentiment analysis / classification (for example grammar rules, which verb/ noun to place emphasis on), those earlier layers should be fine-tuned at a lower learning rate to prevent drastic decreases in accuracy.
How do we go about using these techniques in classifying texts?
We have to save the language model that we trained as an encoder, which means that all layers are saved except the last layer:

We first create a text datablock, before passing it into a dataloader from the dataset, which is slightly different from the one that we saw earlier. The difference between this, and the previous dataloader that we created, is that we are now getting the model to classify using the classic get_x and get_y functions. Furthermore, we are inputting the list of known tokens, or vocabs, into the dataloader as well.

We then create our learner, which utilises the AWD_LSTM framework as shown before. We now train this model for one epoch, establishing a baseline accuracy of 56% for our classifier.
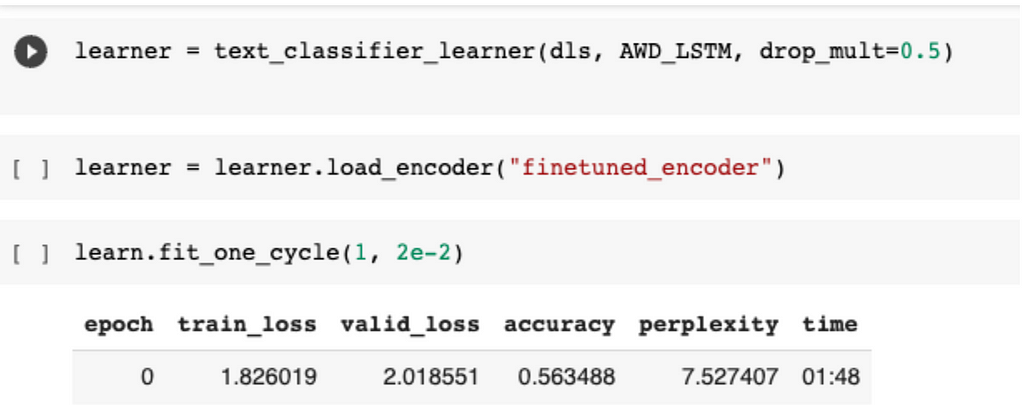
Now here’s the interesting part.
Previously, when I was training my model, I did not utilise the technique of gradual unfreezing and discriminative fine-tuning. What then happened was a constant decrease in accuracy, despite me training it multiple times using a constant learning rate. This was something that I had previously not encountered, and I figured that this would be a good time for me to resolve this problem by trying out the ULMFiT technique. Surprisingly enough, it worked, and accuracy started to increase after several epochs of training.
In this code shown below:
learn.freeze_to(-2)
What we are doing is basically freezing all layers except for the last 2 parameter groups, which is basically the practice of gradual unfreezing.
What is discriminative fine tuning in this case? The answer lies in the next line of code.
learn.fit_one_cycle(1, slice(1e-2/(2.6**4),1e-2))
This line of code basically takes a starting learning rate of (1e-2 divided by 2.6⁴), and has an ending learning rate of 1e-2, with this running for 1 epoch.

We then run these two same lines of code while slowly unfreezing the later layers, and changing the learning rates, resulting in an improvement of about 5% from our initial trained classifier model of one epoch.
We can see the output of our model by calling learner.predict(“”), and then inputting the relevant patient transcript into it.

Some learning points:
Being passionate about the intersection between healthcare and AI is something that has to be cultivated over a long period of time. Personally, that interest grew because I had access to good datasets, be it on Kaggle or on the internet.
However, these datasets have huge limitations — the accuracy of the model and the bias of the data may ultimately render the accuracy of the ML system to be marginally better than flipping a coin in the real world. Realistically, I believe that it would be challenging for a budding ML student to develop his own project to make a huge impact in the world without the model having some inherent limitations. With health data being monopolised by a small number of companies, and with patients concerned about privacy issues, it is getting increasingly difficult to find datasets to train SOTA models on to benefit the wider society.
Enter Nightingale Open Science
Fortunately, we have organisations like Nightingale Open Science who seek to provide access to computational medicine. What stands out to me the most is the guarantee of security and ethics that goes into such a platform — where only non-commercial use of the datasets are allowed, so that the knowledge generated from crunching these datasets can benefit everyone.
Moving forward
I’ll be exploring more of the intersection between AI and healthcare in the future. One of my milestones for the next few months is to explore the datasets that Nightingale provides, as well as to continue writing about my takeaways. One thing that I would like to do is to explore some of Mihaela Van der Schaar’s papers, especially some of her big ideas.
P.S. Utilising the ULMFiT technique through FastAI got me a Bronze Medal in this Kaggle competition — a huge step in my journey to become a Kaggle Expert!
If any part of this article resonated / inspired you, feel free to reach out to me here!!
Predicting Medical Specialities from Transcripts: A Complete Walkthrough using ULMFiT was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
from Towards Data Science - Medium https://ift.tt/u1ij0cl
via RiYo Analytics

No comments