https://ift.tt/3ftW73H How to extract information from a huge corpus of data Photo by Ryunosuke Kikuno on Unsplash How do you extract...
How to extract information from a huge corpus of data

How do you extract information contained in a few sentences out of hundreds of thousands of documents? How do you annotate a huge corpus of text?
In this tutorial, we train and build a named entity recogniser (NER) to detect COVID-19 risk factors using one of the largest medical datasets available.
The dataset: cord-19
The cord-19 dataset has over 500,000 research papers about COVID-19 and other coronavirus-related diseases put together and regularly updated by the AllenAI institute. The data used for this tutorial followed similar processing to what is shown on the github repository for the dataset: https://github.com/allenai/cord19.
Our objective is to find the top risk factors mentioned by medical papers for getting severe COVID.
How do we go on about annotating this?
One issue of working with a very large corpus such as this one, is that the information will only be in a few paragraphs in a few documents out of hundreds of thousands. It’s like looking for a needle in a haystack. How do we annotate such a corpus? The only way is to narrow down the problem to a smaller subset of documents that are likely to contain the information we need. For that, we can use regular expressions or other similar rules to first search likely candidates that have the information we need.
For the purpose of exploring and annotating this dataset, we use open-source library DataQA, a library based on Python and React for data exploration and labelling. This python package includes the elasticsearch text search engine and can easily be installed with pip.
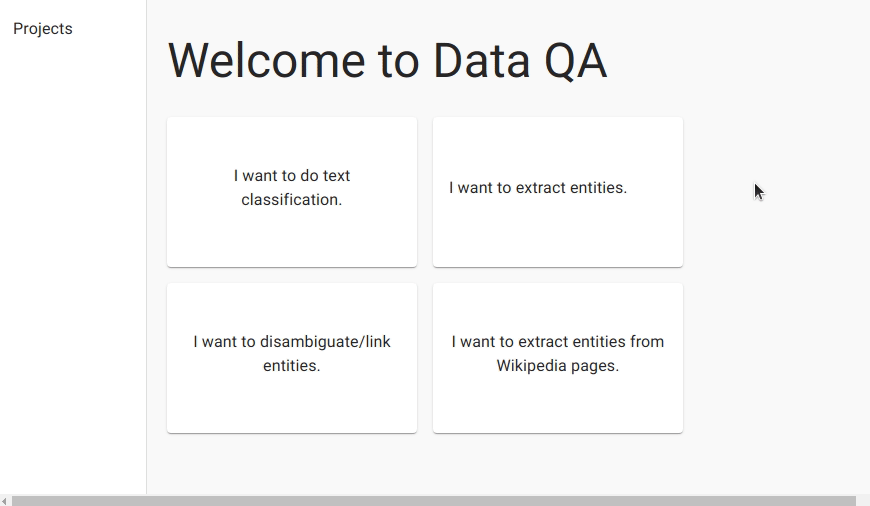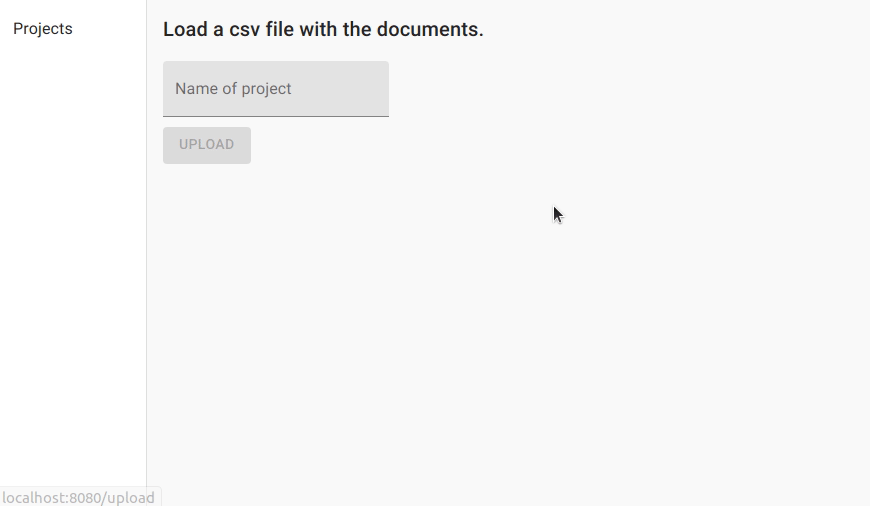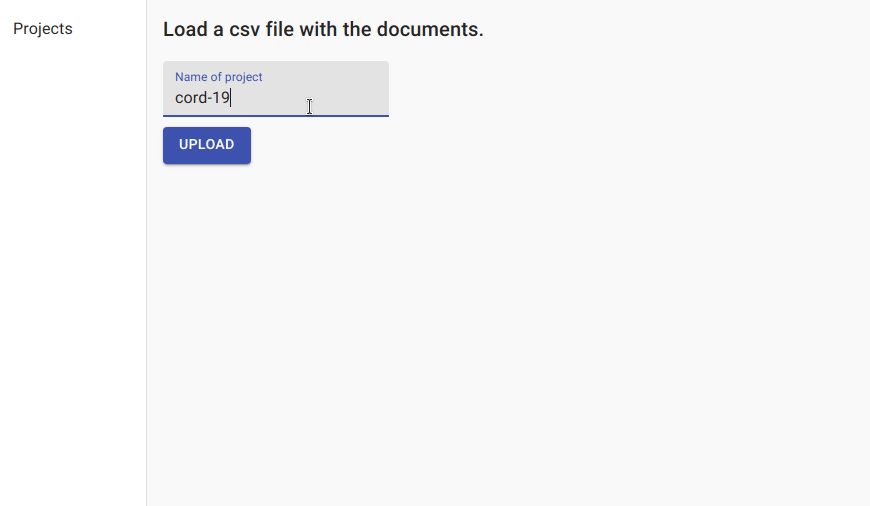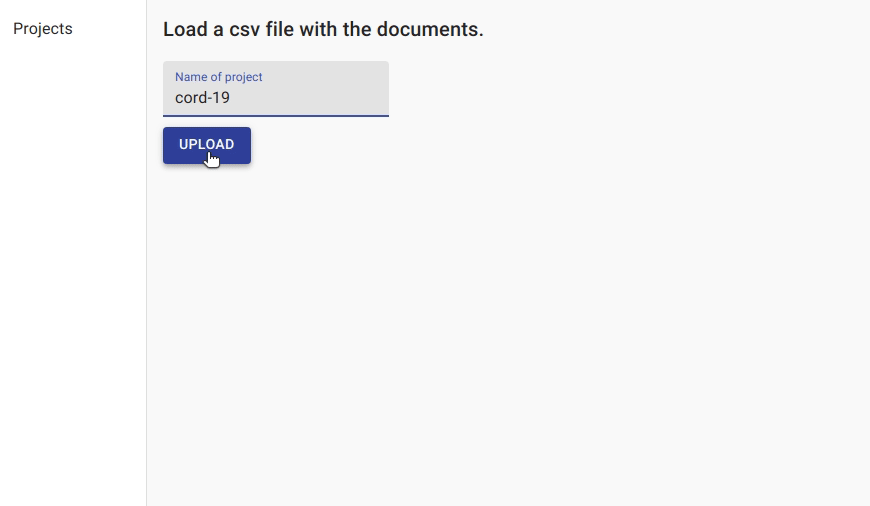
We first create a project for named entity recognition and upload the data.
After our documents have been uploaded and indexed, we need to narrow down on some paragraphs that are likely to contain the information we want. In order to achieve that, we can create our first rule. The rule is as follows: the document must contain a sentence with the word “risk factor” and one of multiple known risk factors, such as “age”, “obesity”, etc.
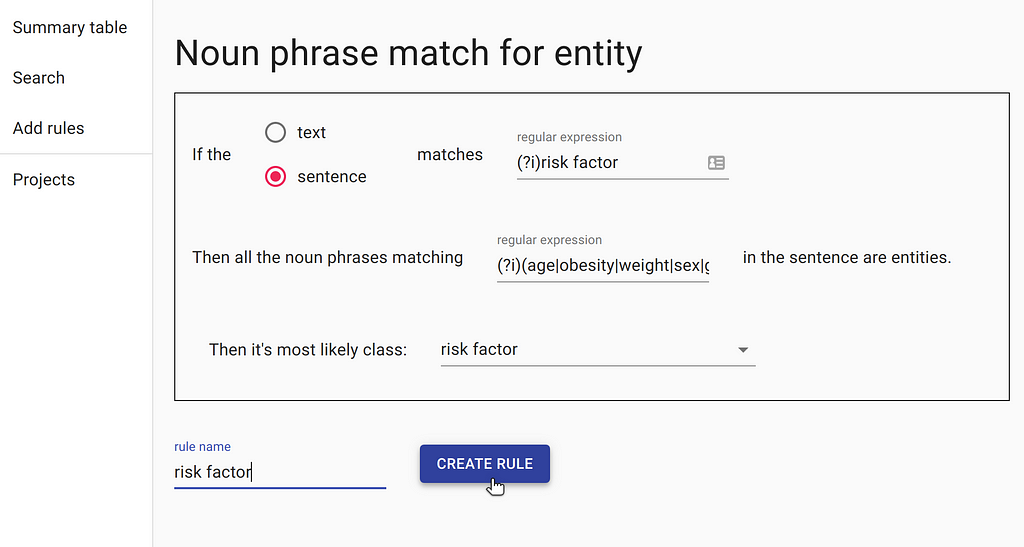
This rule will find the documents that have a sentence summarising all the COVID-19 risk factors known to date. Such a search would not be possible with a standard search engine, that mainly focuses on keyword search. DataQA will also pre-label all the known risk factors using rules, making the manual process more efficient.
We include another rule which is a slightly different way of expressing the same idea: the document must contain a sentence with either “risk” followed by “hospitalisation/hospitalization” (or in reverse order), followed by one of the known risk factors.
Once we have these rules in place, we can either annotate the documents with the pre-labels from the rules or go to the search. In the search, we apply the rules as filters and search for mentions of covid, as there are many papers that discuss risk factors for other non-relevant conditions.
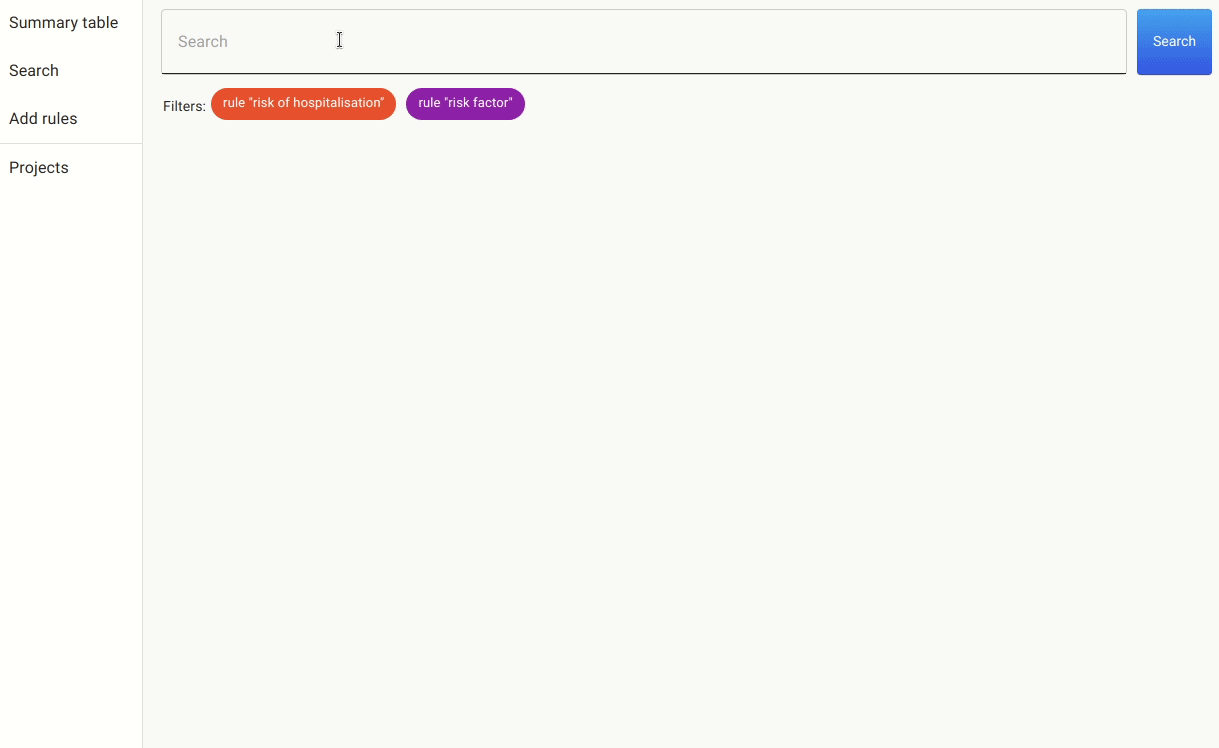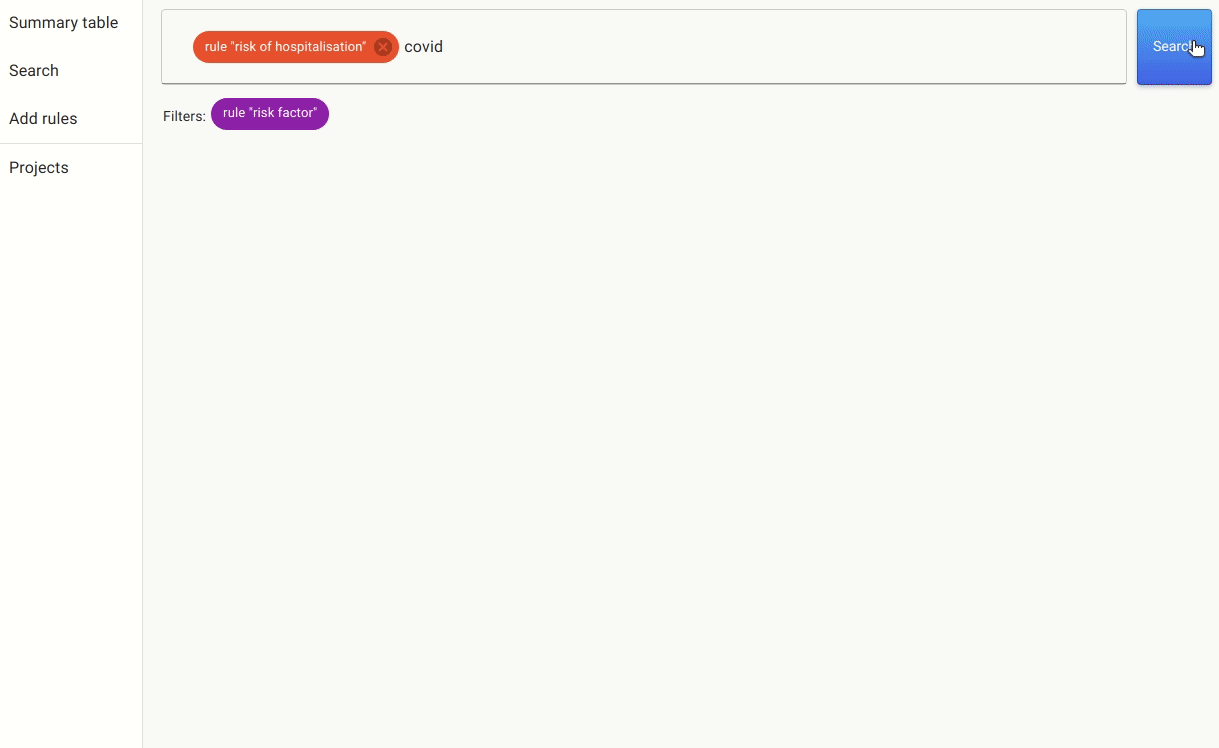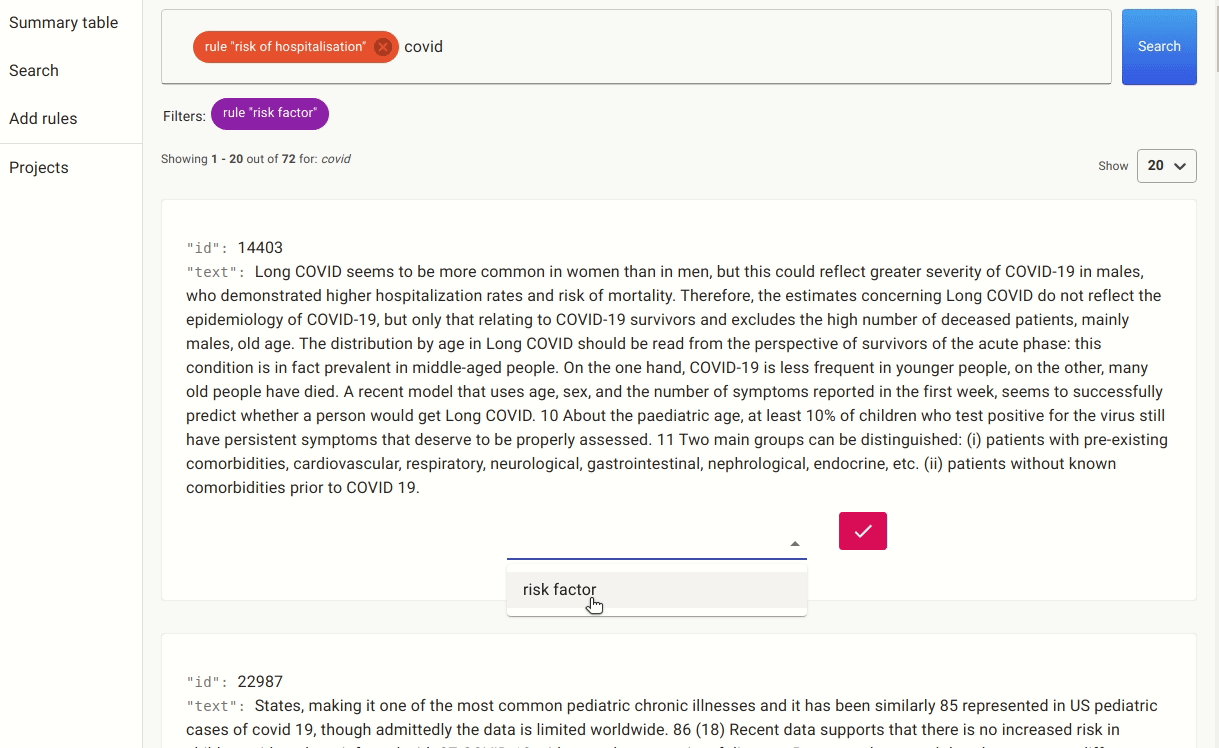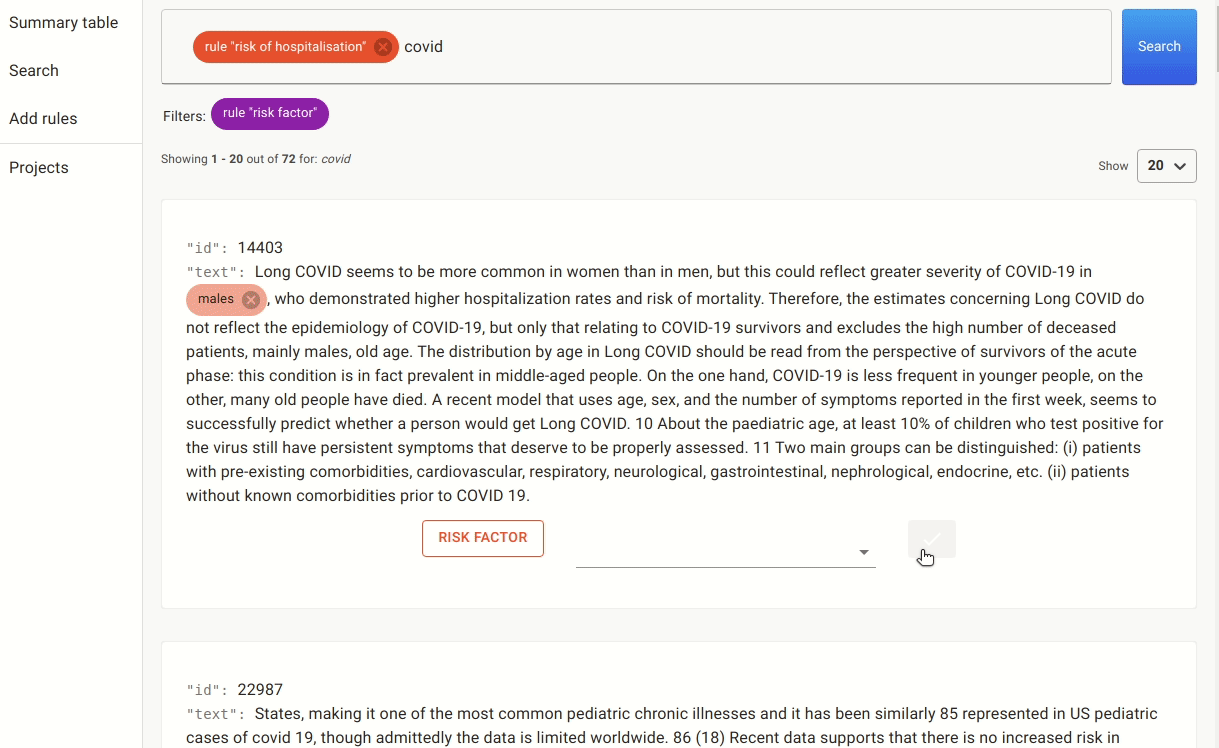
After doing this for a while, we ended up with a small labelled set of 42 documents which we used to train the named entity extractor. It’s a small training dataset, and our goal is to see if we can quickly train a NER to detect more risk factors without the need to do any additional manual labelling.
Building the NER with Spacy
We use Spacy to build a named entity extractor based on the data we just annotated. We first need to prepare the training data.
Then, the code to train our NER is as follows. We use a Spacy pipeline, adding a sentence segmenter to detect sentence boundaries, and replace the default NER step with a new one that only has “risk factor” as an entity. When then train the pipeline in mini-batches to avoid overfitting.
Finding new mentions of risk factors
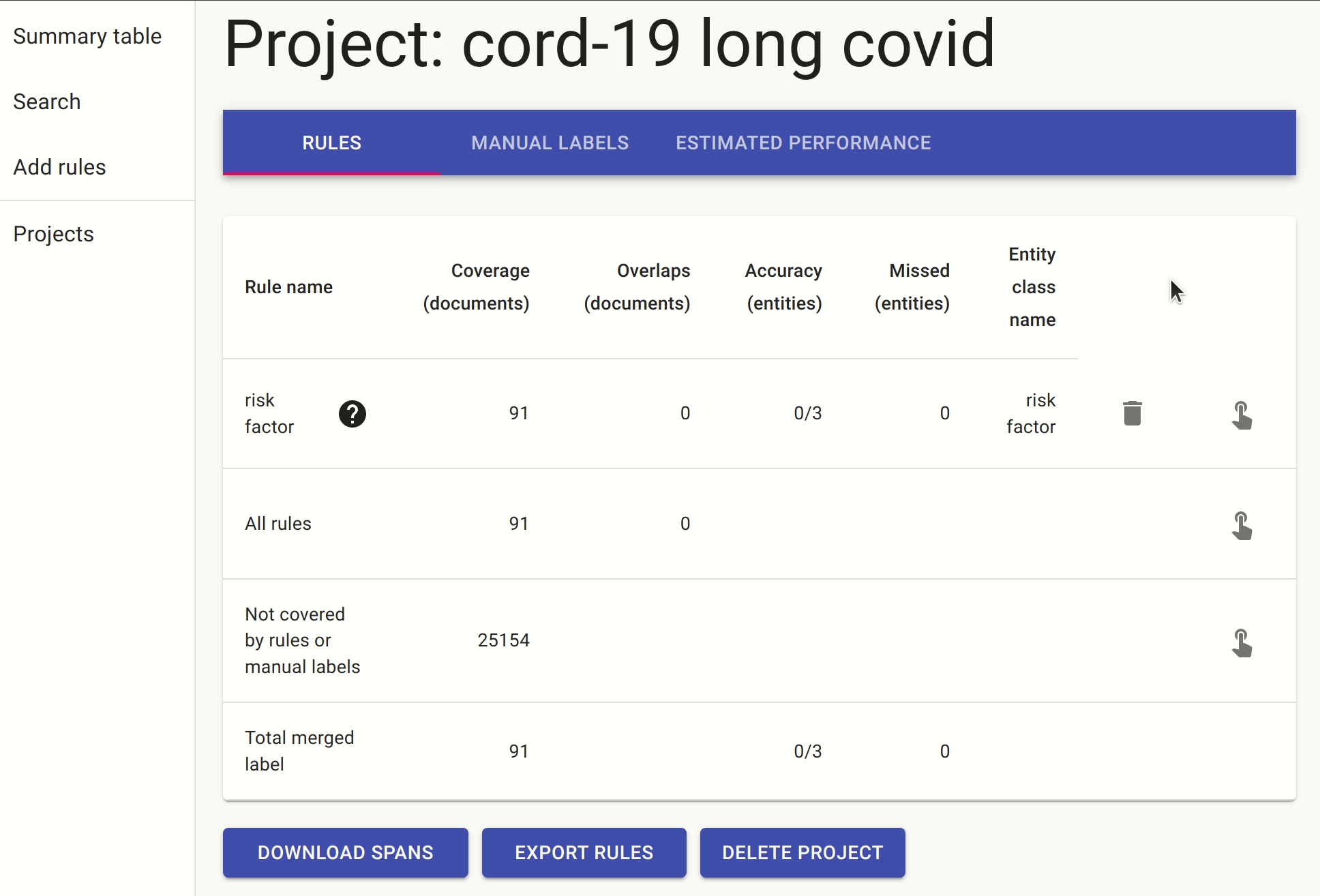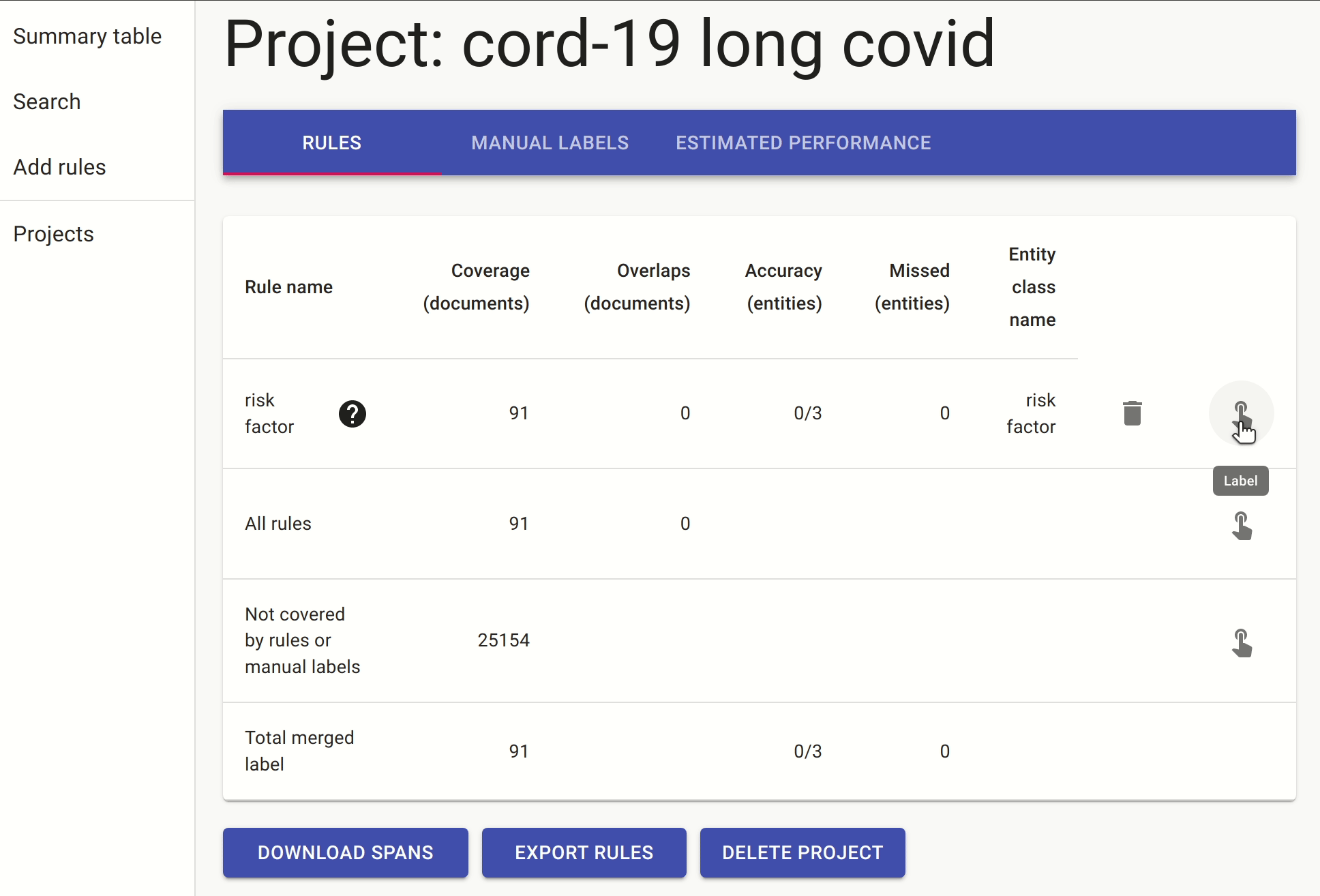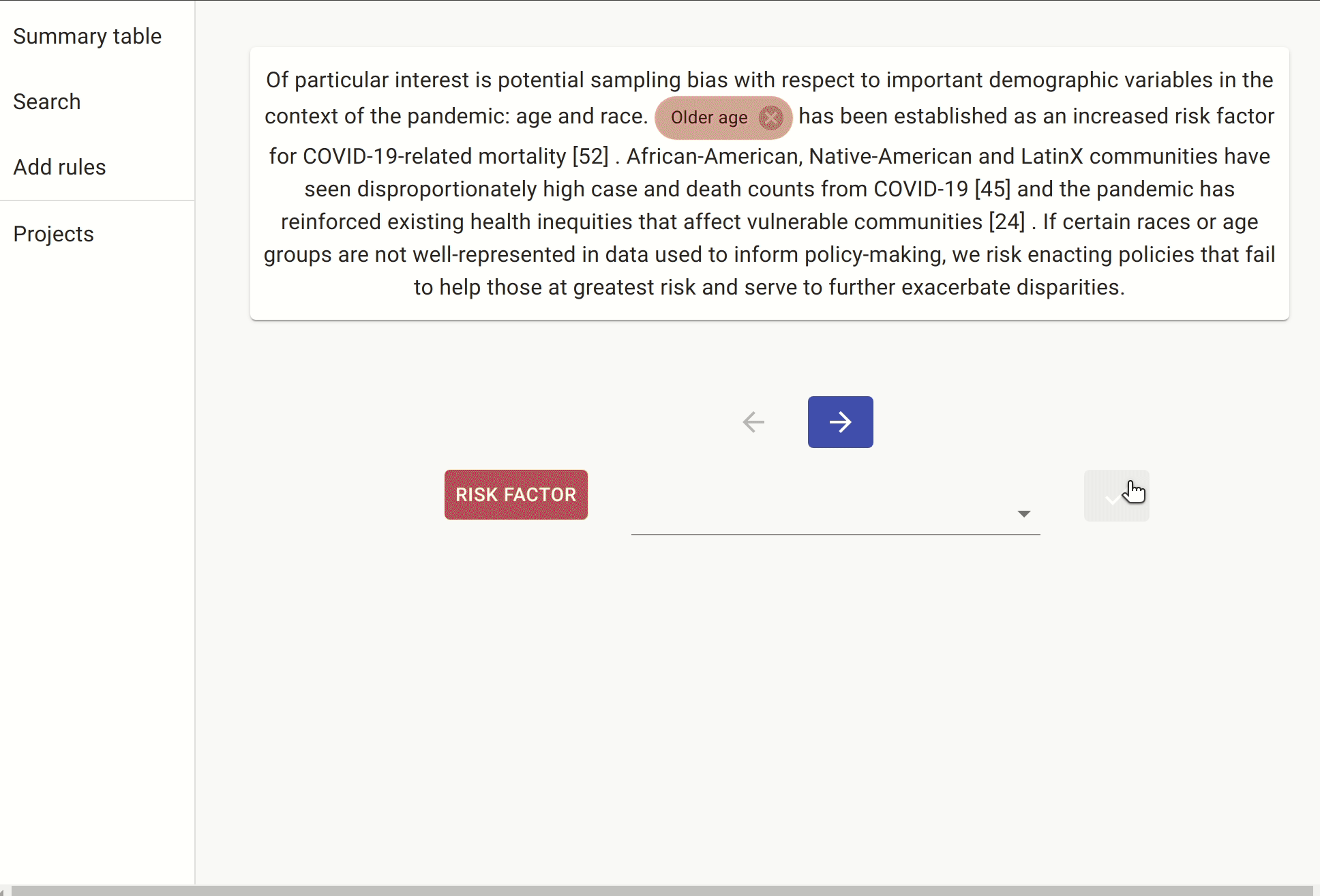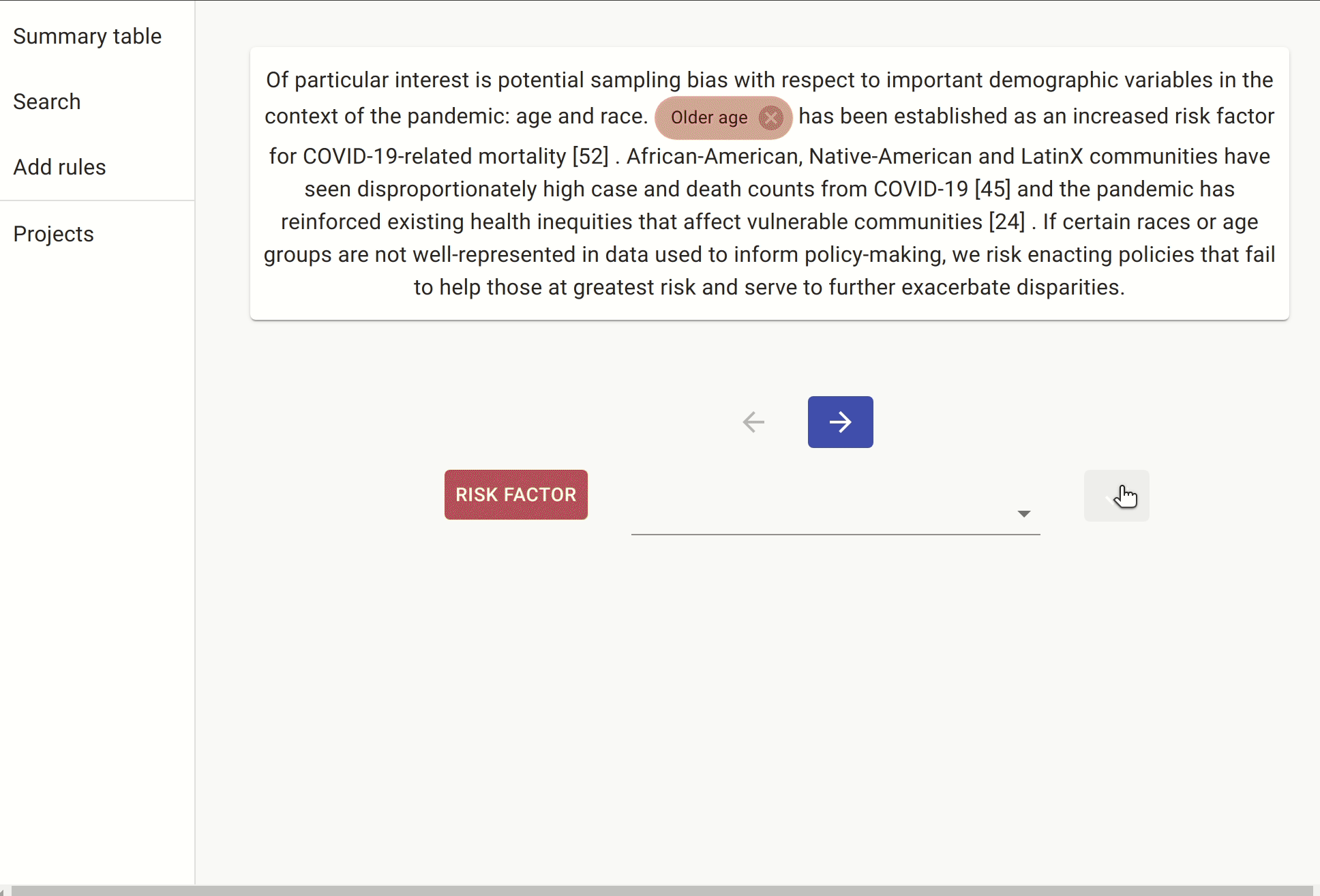
Once we have trained our NER, we can then run it over a sample of the unlabelled set of documents to see if we can find a new set of risk factors we had not identified during the manual labelling process. Something worth mentioning here is that we are running our model over the documents singled out by the rules above, so as to avoid getting too many false positives.
Here are some examples of risk factors identified by the NER that had not been labelled manually: neuromuscular disorders, race/ethnicity, prepregnancy obesity, etc.
All in all, putting all our findings together, the top 20 risk factors mentioned in the literature for severe covid ranked by the number of papers mentioning them in the sample were:
[('diabetes', 87),
('hypertension', 84),
('age', 79),
('obesity', 67),
('smoking', 36),
('sex', 27),
('cardiovascular disease', 27),
('gender', 27),
('older age', 20),
('copd', 19),
('asthma', 18),
('male sex', 18),
('chronic kidney disease', 16),
('chronic obstructive pulmonary disease', 14),
('bmi', 13),
('pneumonia', 13),
('therapy', 12),
('cardiovascular', 11),
('cancer', 11),
('diabetes mellitus', 11)]
Some of the risk factors point to the same underlying entity (e.g. “age” and “older age” are the same risk factor) and “therapy” is a false position that does not belong to the list. However, this list gives us some insight into the key factors discussed in the literature.
What about long covid?
There are fewer papers talking about risk factors for long COVID given the scarcity of data. Searching for those mentions is thus challenging because there is little information.
We use the same approach we took as before to filter on the likely candidates. This time, the number of papers was much smaller, and so we could simply get the information directly.
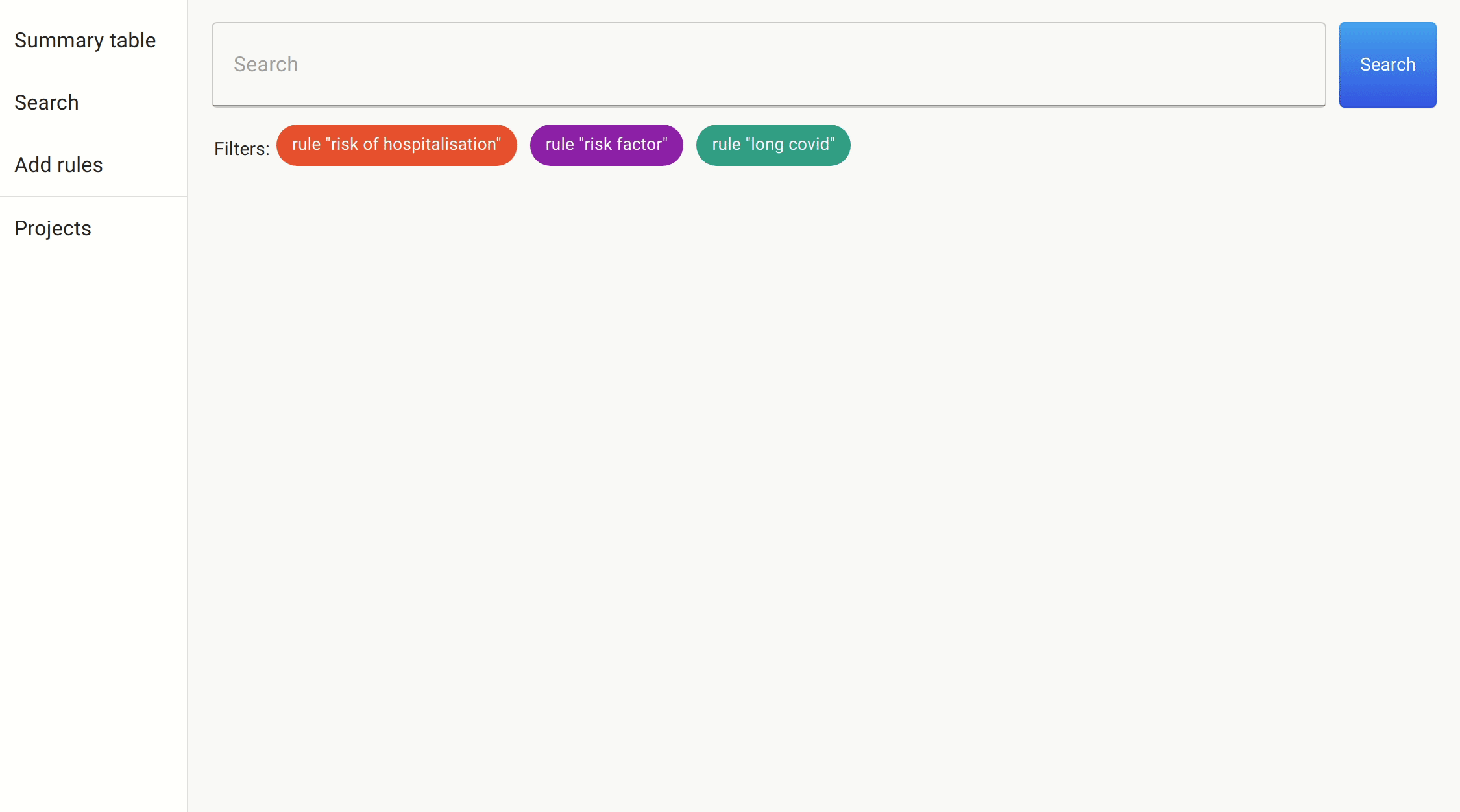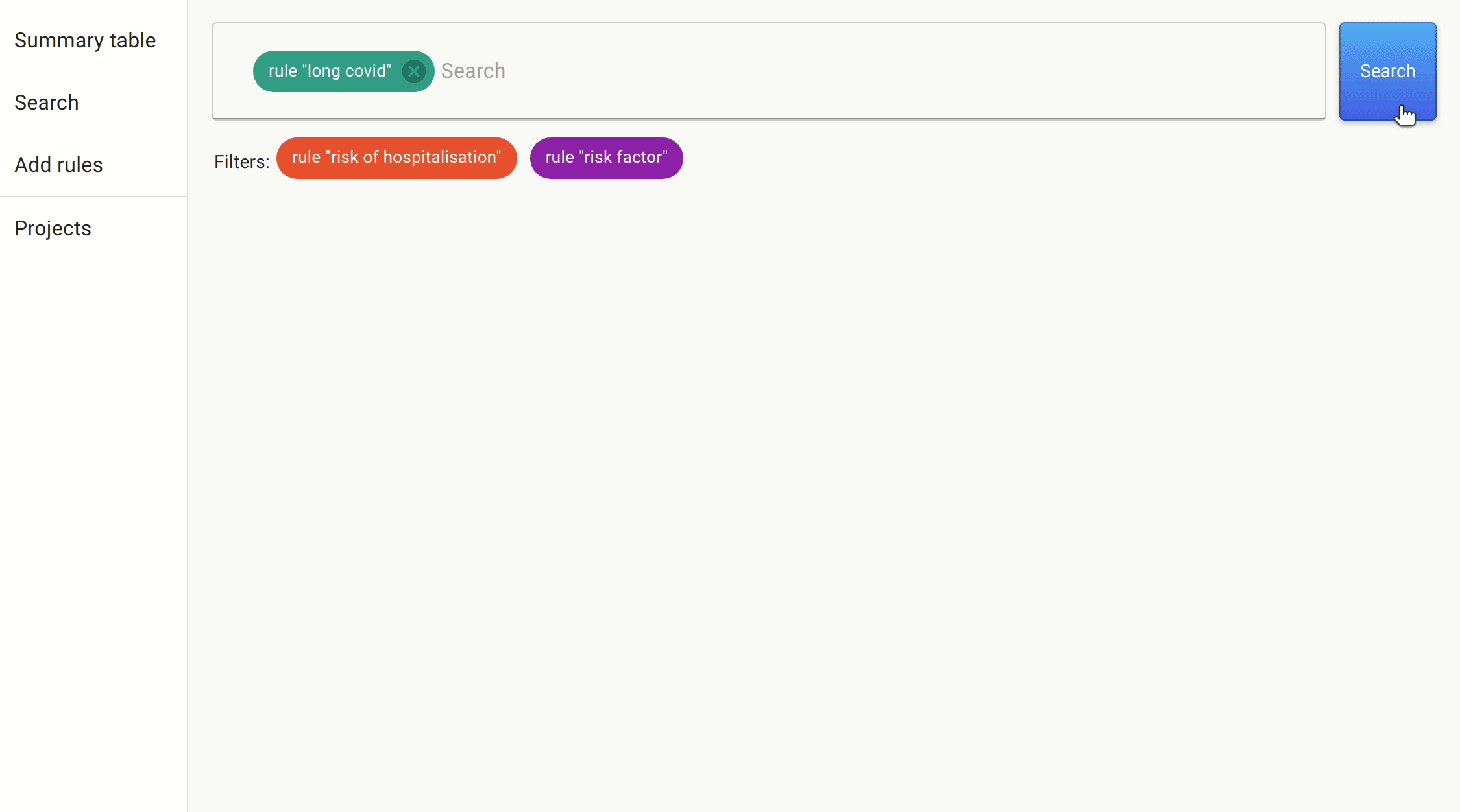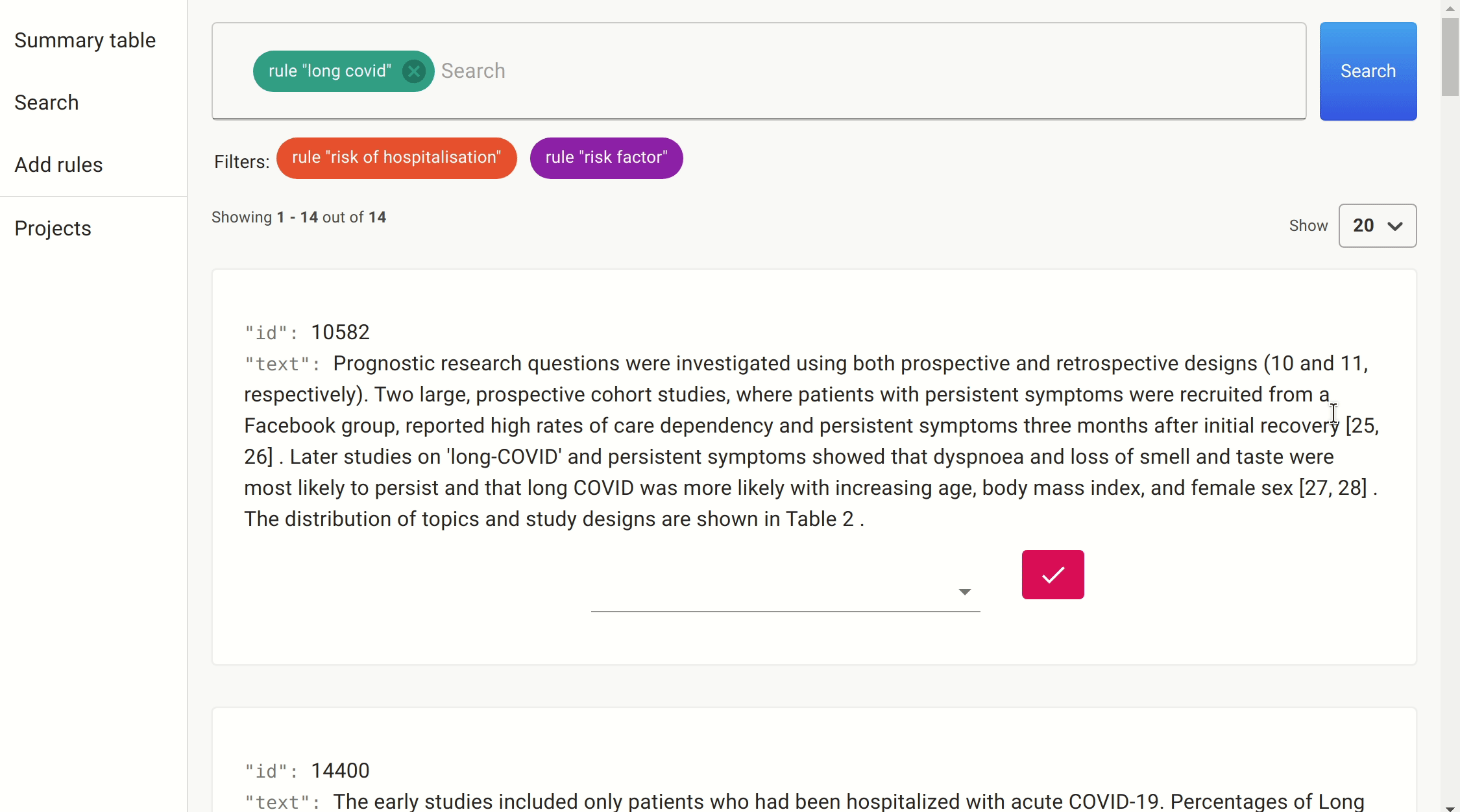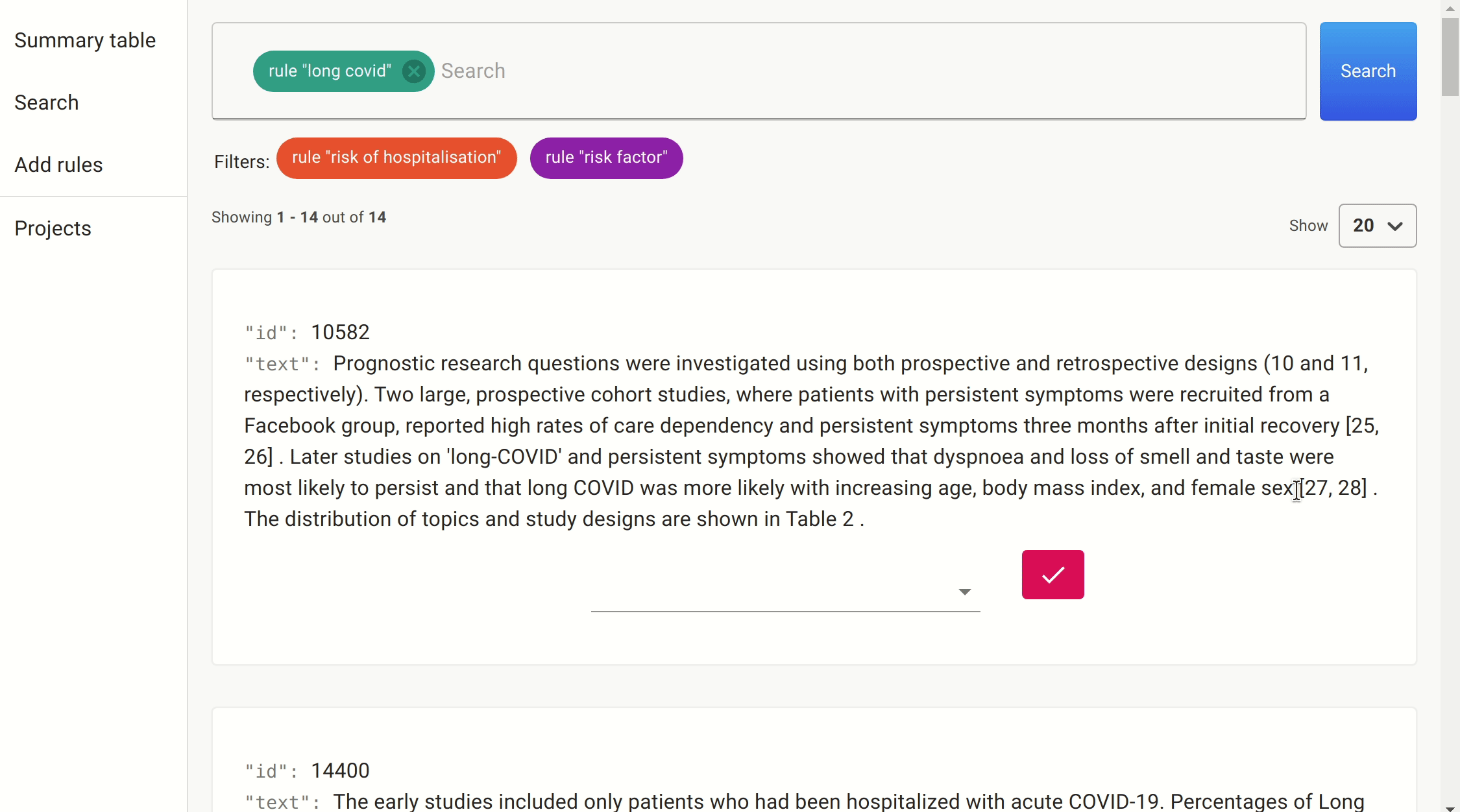
After a quick search, we found a number of papers citing female sex, increasing age, increasing BMI, and number of symptoms reported in the first week as predictive features of whether someone with COVID-19 would develop long COVID.
If you would like to find out more about data exploration with DataQA, head to our repository for more tutorials and examples. We also offer an enterprise version of the product that works directly with pdfs (check our website). Do not forget to leave a star to support this open-source project :-)
Building a named entity extractor for COVID-19 risk factors was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
from Towards Data Science - Medium https://ift.tt/3KjQP91
via RiYo Analytics

ليست هناك تعليقات