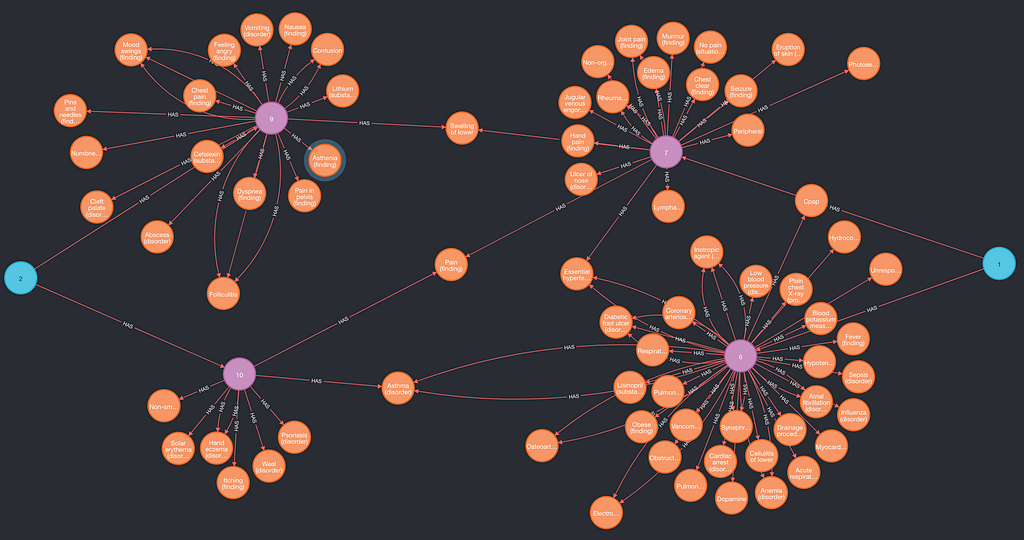
https://ift.tt/303Siy1 Figure 1: Diseases, Symptoms, Medications and Procedures associated with two Patients(blue) via Documents (Pink). I...
A hands-on tutorial on making biomedical concepts extracted from free text easily accessible to clinicians and researchers
Biomedical NER+L is concerned with extracting concepts from free text found in Electronic Health Records (EHRs) and linking them to large biomedical databases like SNOMED-CT and UMLS.
In this post, we will focus on what to do once concepts are extracted from free text, in other words, how to make them useful to clinicians and other researchers.
Prerequisites: None, unless you want to replicate the results then you need to be able to use MedCAT and Neo4j.
Introduction
Assume we got access to a hospital and annotated all the free text in Electronic Health Records for SNOMED concepts. If we have used MedCAT then the output is stored into a.json file. Our aim is to move the extracted concepts into a database that will allow anyone to write simple queries and utilize the annotations, e.g.:
- Return all patients that have diabetes and leg pain
- Return all diseases found for patient X or Return all diseases found in a specific document
- Return all patients that have the symptom vomiting and are taking the drug valpam
- Return all male patients that do not have Alzheimer's but have had two or more mentions of seizure.
- Return all patients that have Dementia or any other disease that is a direct child of the concept Dementia in the SNOMED ontology.
While a relational database could do the job (with some twists and turns), a graph database is better suited. Why? The first reason is that we want to have the SNOMED/UMLS ontology (basically a ~directed graph) imported so that we can easily fulfil the last query from the examples above. And second, our annotations can be easily represented as a graph: Patient-[HAS]->Document-[HAS]->Concept. In words: a patient can have one or many documents, a document can have one or many mentions of a concept (e.g. diseases).
The rest of the post will be, to some extent, a tutorial (accompanied by a Jupyter Notebook). It assumes that you have a Neo4j database installed and available on localhost:7474. As we cannot use real data, I’ve created two dummy files from mtsamples:
- patients.csv contains basic information about all patients
- documents.csv contains the document text + a document ID
Annotating documents
Before we do anything we need to generate the annotations for the free text available in documents.csv if you already have documents processed with MedCAT or any other NER+L tool, then you can skip this step.
from medcat.cat import CAT
df_docs = pd.read_csv('./documents.csv')
# This would be a generator if we have a lot of docs
data = [(k,v) for k,v in df_docs[['documentId', 'text']].values]
# You can find the model in the medcat repository
cat = CAT.load_model_pack('./medmen_wstatus_2021_oct.zip')
docs = cat.multiprocessing(data, nproc=10)
json.dump(docs, open("./annotations.csv", 'w'))
Populating Neo4j
Create indexes to speed up data ingestion and search (as the dataset can be very large)
from medcat.neo.data_preparation import *
from medcat.neo.neo_connector import NeoConnector
import pandas as pd
# Helper for sending requests to neo
neo = NeoConnector('bolt://localhost:7687/', user='neo4j')
# Indexes are pre-defined in the data_preparation helper
for ind in get_index_queries():
try:
neo.execute(ind)
except Exception as e:
print(e)
Next, start the imports.
# Import Patients
df_pts = pd.read_csv('./patients.csv')
# If you cannot write to this output_dir save somewhere else and copy
q = create_patients_csv(df_pts, output_dir='/var/lib/neo4j/import/')
# Run the query for import
neo.execute(q)
In the neo4j browser (localhost:7474) there will now be 182 new nodes with the label Patient. The same will be repeated for Concepts, Documents and finally Annotations. Details are in the accompanying Jupyter Notebook, here we are skipping and assuming everything is imported.
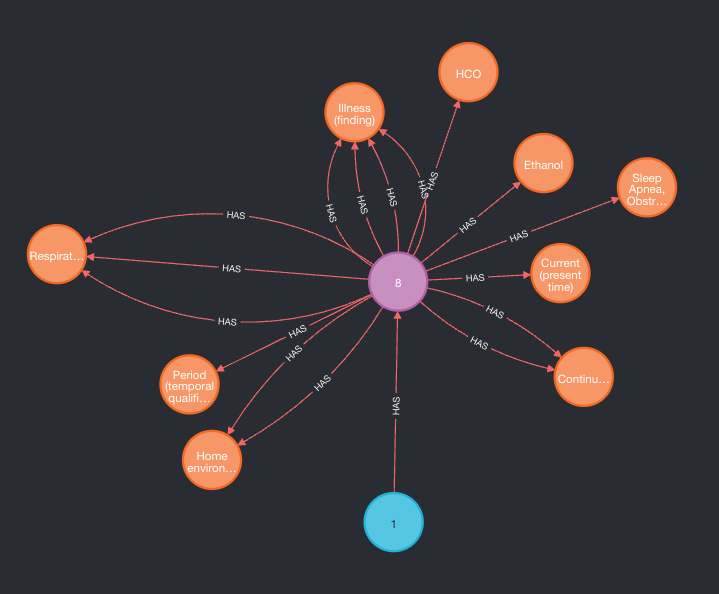
The following query should now work and return a sample of the data:
MATCH (p:Patient)-->(d:Document)-->(c:Concept) RETURN p, d, c LIMIT 10
The output graph(differences are possible, important is to get back all three node types):
Querying Neo4j
We can now easily translate each one of the natural language queries we’ve shown above into Cypher (the neo4j query language). But, if we do not know Cypher, MedCAT has a small class that can be used to query Neo4j:
# Get all patients that have the concepts (Fever and Sleep Apnea). #We will set the ignore_meta flag as the medmentions model does not
#have all required meta annotations.
patients, q = neo.get_all_patients(concepts=['C0520679', 'C0015967'], limit=10, ignore_meta=True)
Or to get all diseases assigned to a patient or document:
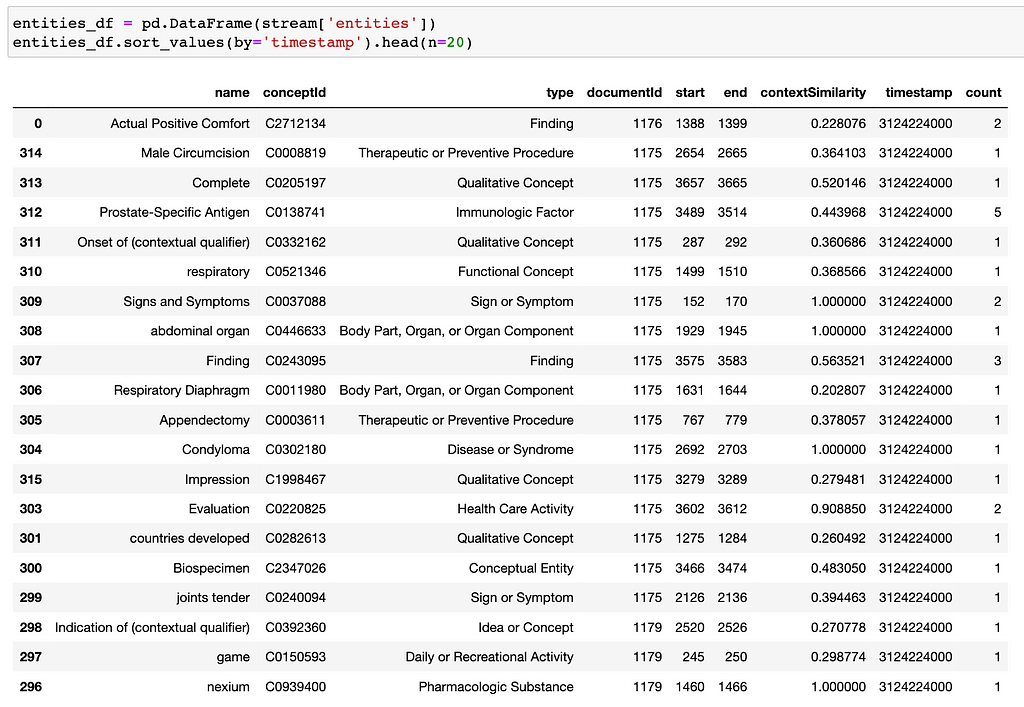
# Get all concepts from one patient
stream, q = neo.get_all_concepts_from(patient_id='32', bucket_size_seconds=10**10)
The results can be displayed as a Dataframe:
End
That’s it, the medcat.neo library will be updated over time with new queries and functions for import/export.
Thank you for reading.
Exploring Electronic Health Records with MedCAT and Neo4j was originally published in Towards Data Science on Medium, where people are continuing the conversation by highlighting and responding to this story.
from Towards Data Science - Medium https://ift.tt/3drG45t
via RiYo Analytics


No comments